Lahsen Boulmane
Hierarchical ResNeXt Models for Breast Cancer Histology Image Classification
Oct 21, 2018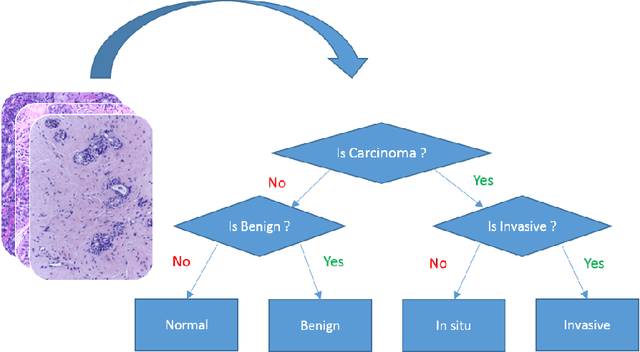
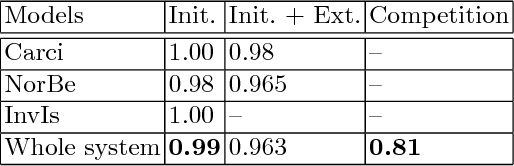
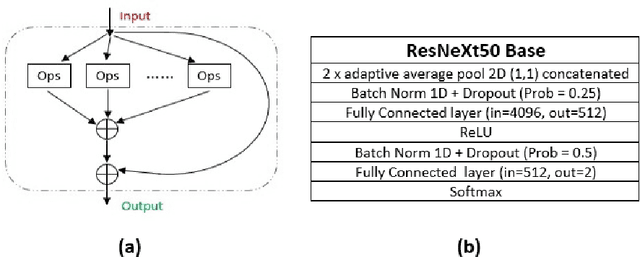
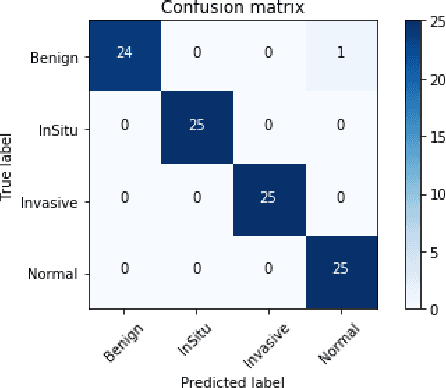
Abstract:Microscopic histology image analysis is a cornerstone in early detection of breast cancer. However these images are very large and manual analysis is error prone and very time consuming. Thus automating this process is in high demand. We proposed a hierarchical system of convolutional neural networks (CNN) that classifies automatically patches of these images into four pathologies: normal, benign, in situ carcinoma and invasive carcinoma. We evaluated our system on the BACH challenge dataset of image-wise classification and a small dataset that we used to extend it. Using a train/test split of 75%/25%, we achieved an accuracy rate of 0.99 on the test split for the BACH dataset and 0.96 on that of the extension. On the test of the BACH challenge, we've reached an accuracy of 0.81 which rank us to the 8th out of 51 teams.
BACH: Grand Challenge on Breast Cancer Histology Images
Aug 13, 2018
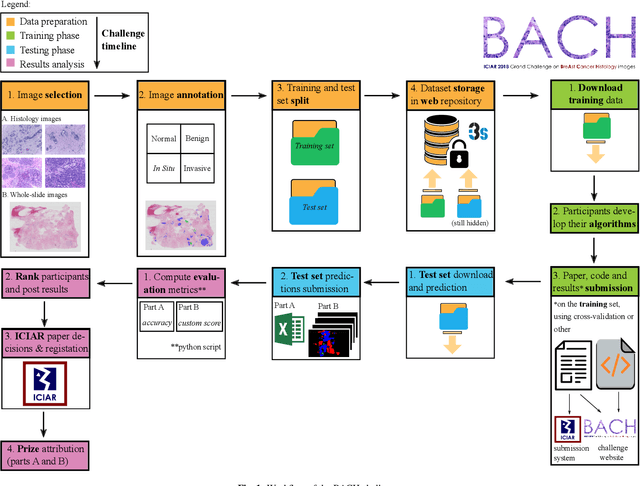
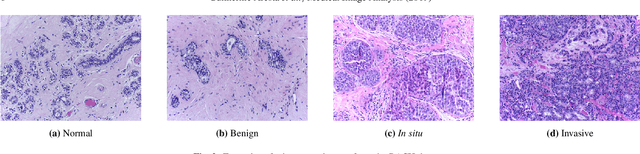
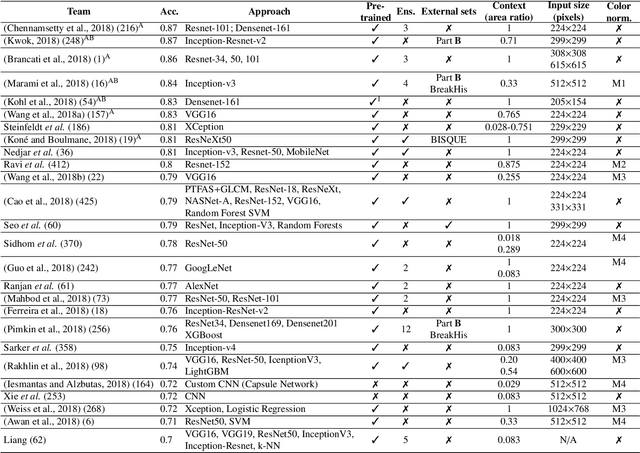
Abstract:Breast cancer is the most common invasive cancer in women, affecting more than 10% of women worldwide. Microscopic analysis of a biopsy remains one of the most important methods to diagnose the type of breast cancer. This requires specialized analysis by pathologists, in a task that i) is highly time- and cost-consuming and ii) often leads to nonconsensual results. The relevance and potential of automatic classification algorithms using hematoxylin-eosin stained histopathological images has already been demonstrated, but the reported results are still sub-optimal for clinical use. With the goal of advancing the state-of-the-art in automatic classification, the Grand Challenge on BreAst Cancer Histology images (BACH) was organized in conjunction with the 15th International Conference on Image Analysis and Recognition (ICIAR 2018). A large annotated dataset, composed of both microscopy and whole-slide images, was specifically compiled and made publicly available for the BACH challenge. Following a positive response from the scientific community, a total of 64 submissions, out of 677 registrations, effectively entered the competition. From the submitted algorithms it was possible to push forward the state-of-the-art in terms of accuracy (87%) in automatic classification of breast cancer with histopathological images. Convolutional neuronal networks were the most successful methodology in the BACH challenge. Detailed analysis of the collective results allowed the identification of remaining challenges in the field and recommendations for future developments. The BACH dataset remains publically available as to promote further improvements to the field of automatic classification in digital pathology.
Hybrid Forests for Left Ventricle Segmentation using only the first slice label
Apr 30, 2018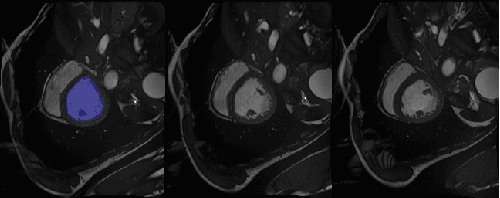

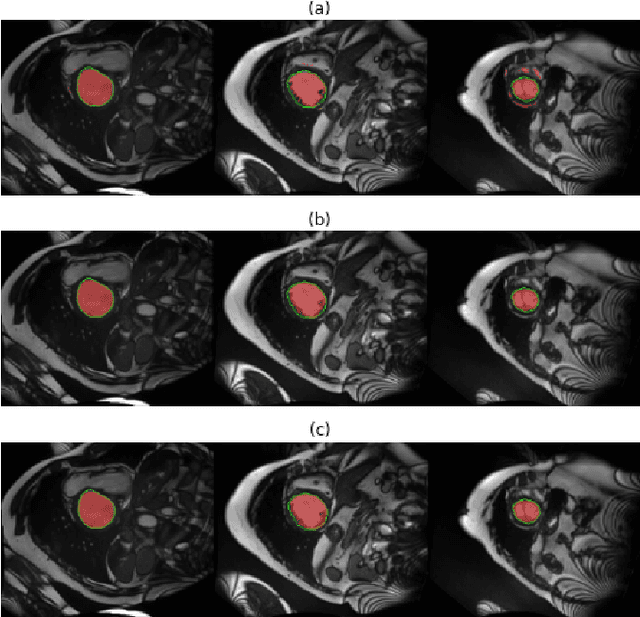
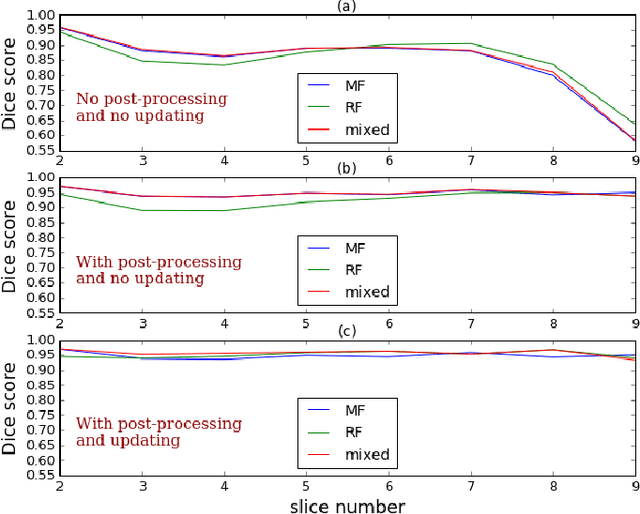
Abstract:Machine learning models produce state-of-the-art results in many MRI images segmentation. However, most of these models are trained on very large datasets which come from experts manual labeling. This labeling process is very time consuming and costs experts work. Therefore finding a way to reduce this cost is on high demand. In this paper, we propose a segmentation method which exploits MRI images sequential structure to nearly drop out this labeling task. Only the first slice needs to be manually labeled to train the model which then infers the next slice's segmentation. Inference result is another datum used to train the model again. The updated model then infers the third slice and the same process is carried out until the last slice. The proposed model is an combination of two Random Forest algorithms: the classical one and a recent one namely Mondrian Forests. We applied our method on human left ventricle segmentation and results are very promising. This method can also be used to generate labels.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge