Julien Fauqueur
Sparks of Tabular Reasoning via Text2SQL Reinforcement Learning
Apr 23, 2025



Abstract:This work reframes the Text-to-SQL task as a pathway for teaching large language models (LLMs) to reason over and manipulate tabular data--moving beyond the traditional focus on query generation. We propose a two-stage framework that leverages SQL supervision to develop transferable table reasoning capabilities. First, we synthesize detailed chain-of-thought (CoT) traces from real-world SQL queries, providing step-by-step, clause-level supervision that teaches the model how to traverse, filter, and aggregate table fields. Second, we introduce a Group Relative Policy Optimization (GRPO) reinforcement learning objective that connects SQL execution accuracy to generalizable reasoning by encouraging steps that extend beyond task-specific syntax and transfer across datasets. Empirically, our approach improves performance on standard Text-to-SQL benchmarks and achieves substantial gains on reasoning-intensive datasets such as BIRD and CRT-QA, demonstrating enhanced generalization and interpretability. Specifically, the distilled-quantized LLaMA model achieved a 20\% increase in accuracy when trained on Text-to-SQL tasks, while Qwen achieved a 5\% increase. These results suggest that SQL can serve not only as a target formalism but also as an effective scaffold for learning robust, transferable reasoning over structured data.
Learning Informative Representations of Biomedical Relations with Latent Variable Models
Nov 20, 2020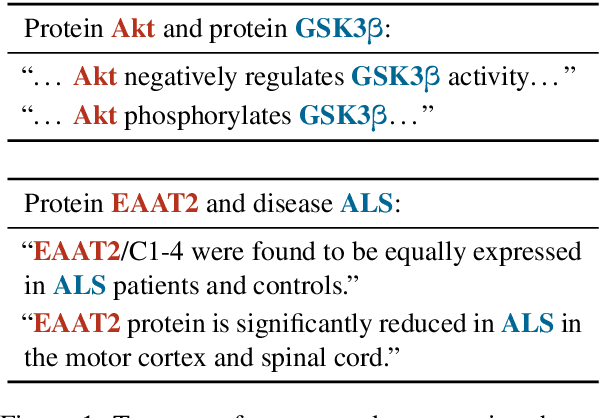

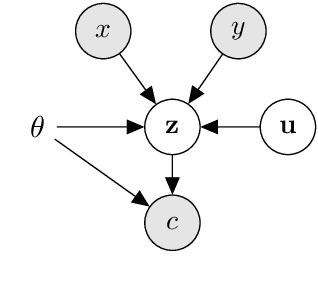
Abstract:Extracting biomedical relations from large corpora of scientific documents is a challenging natural language processing task. Existing approaches usually focus on identifying a relation either in a single sentence (mention-level) or across an entire corpus (pair-level). In both cases, recent methods have achieved strong results by learning a point estimate to represent the relation; this is then used as the input to a relation classifier. However, the relation expressed in text between a pair of biomedical entities is often more complex than can be captured by a point estimate. To address this issue, we propose a latent variable model with an arbitrarily flexible distribution to represent the relation between an entity pair. Additionally, our model provides a unified architecture for both mention-level and pair-level relation extraction. We demonstrate that our model achieves results competitive with strong baselines for both tasks while having fewer parameters and being significantly faster to train. We make our code publicly available.
Constructing large scale biomedical knowledge bases from scratch with rapid annotation of interpretable patterns
Jul 03, 2019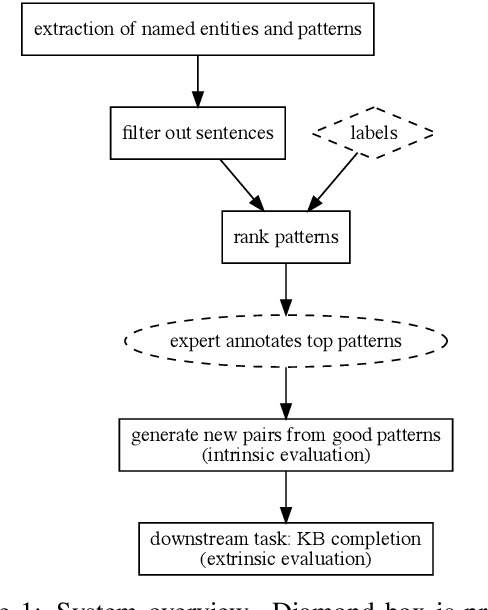
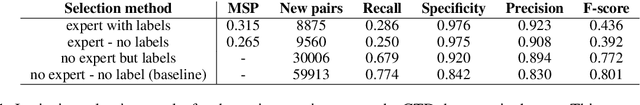
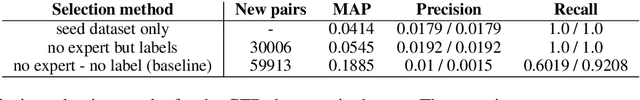
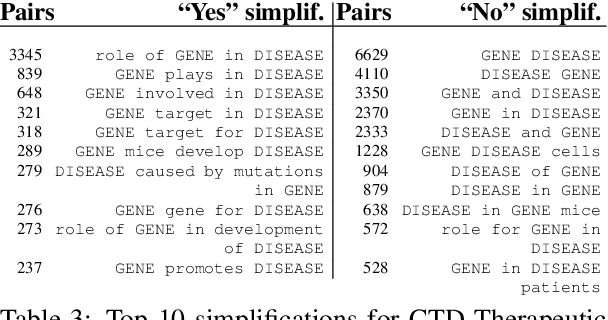
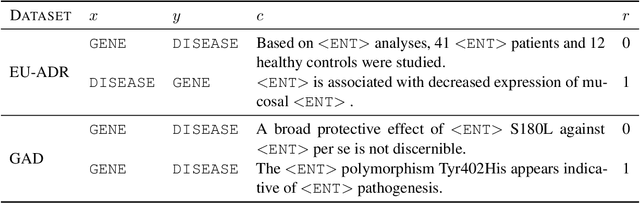
Abstract:Knowledge base construction is crucial for summarising, understanding and inferring relationships between biomedical entities. However, for many practical applications such as drug discovery, the scarcity of relevant facts (e.g. gene X is therapeutic target for disease Y) severely limits a domain expert's ability to create a usable knowledge base, either directly or by training a relation extraction model. In this paper, we present a simple and effective method of extracting new facts with a pre-specified binary relationship type from the biomedical literature, without requiring any training data or hand-crafted rules. Our system discovers, ranks and presents the most salient patterns to domain experts in an interpretable form. By marking patterns as compatible with the desired relationship type, experts indirectly batch-annotate candidate pairs whose relationship is expressed with such patterns in the literature. Even with a complete absence of seed data, experts are able to discover thousands of high-quality pairs with the desired relationship within minutes. When a small number of relevant pairs do exist - even when their relationship is more general (e.g. gene X is biologically associated with disease Y) than the relationship of interest - our system leverages them in order to i) learn a better ranking of the patterns to be annotated or ii) generate weakly labelled pairs in a fully automated manner. We evaluate our method both intrinsically and via a downstream knowledge base completion task, and show that it is an effective way of constructing knowledge bases when few or no relevant facts are already available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge