Juan M. Gorriz
Uncertainty-driven ensembles of deep architectures for multiclass classification. Application to COVID-19 diagnosis in chest X-ray images
Nov 27, 2020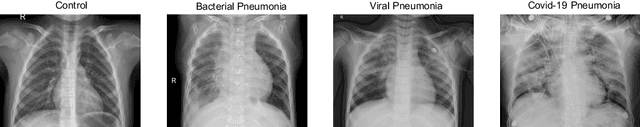
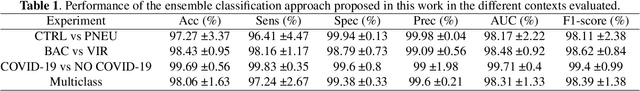
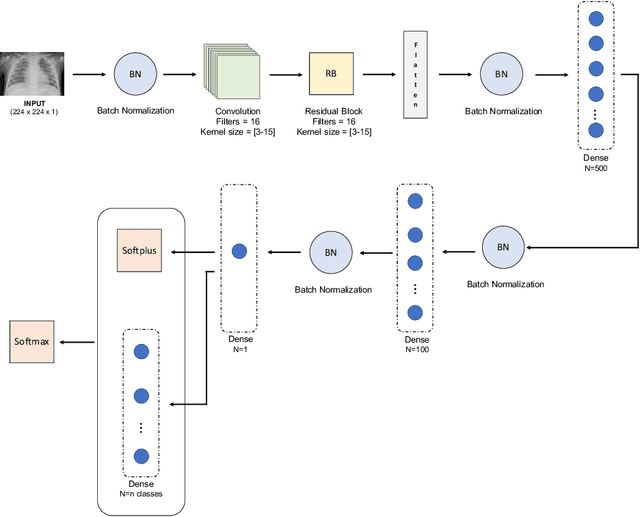
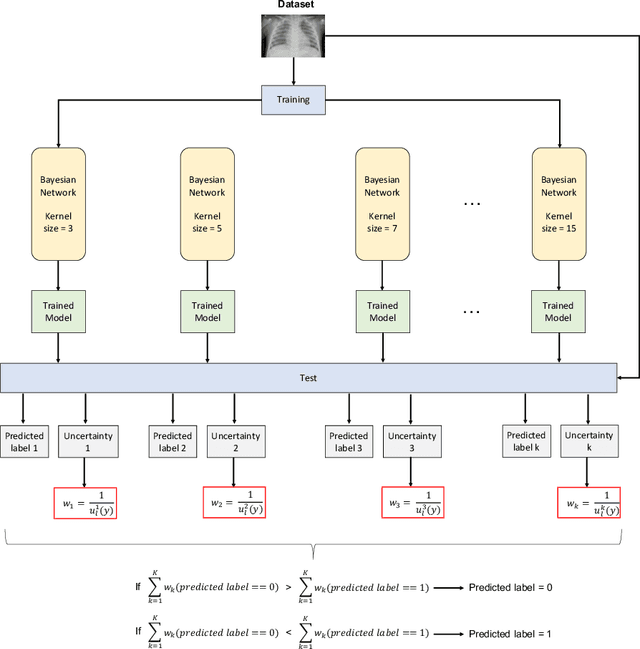
Abstract:Respiratory diseases kill million of people each year. Diagnosis of these pathologies is a manual, time-consuming process that has inter and intra-observer variability, delaying diagnosis and treatment. The recent COVID-19 pandemic has demonstrated the need of developing systems to automatize the diagnosis of pneumonia, whilst Convolutional Neural Network (CNNs) have proved to be an excellent option for the automatic classification of medical images. However, given the need of providing a confidence classification in this context it is crucial to quantify the reliability of the model's predictions. In this work, we propose a multi-level ensemble classification system based on a Bayesian Deep Learning approach in order to maximize performance while quantifying the uncertainty of each classification decision. This tool combines the information extracted from different architectures by weighting their results according to the uncertainty of their predictions. Performance of the Bayesian network is evaluated in a real scenario where simultaneously differentiating between four different pathologies: control vs bacterial pneumonia vs viral pneumonia vs COVID-19 pneumonia. A three-level decision tree is employed to divide the 4-class classification into three binary classifications, yielding an accuracy of 98.06% and overcoming the results obtained by recent literature. The reduced preprocessing needed for obtaining this high performance, in addition to the information provided about the reliability of the predictions evidence the applicability of the system to be used as an aid for clinicians.
Automated detection and segmentation of non-mass enhancing breast tumors with dynamic contrast-enhanced magnetic resonance imaging
Sep 26, 2018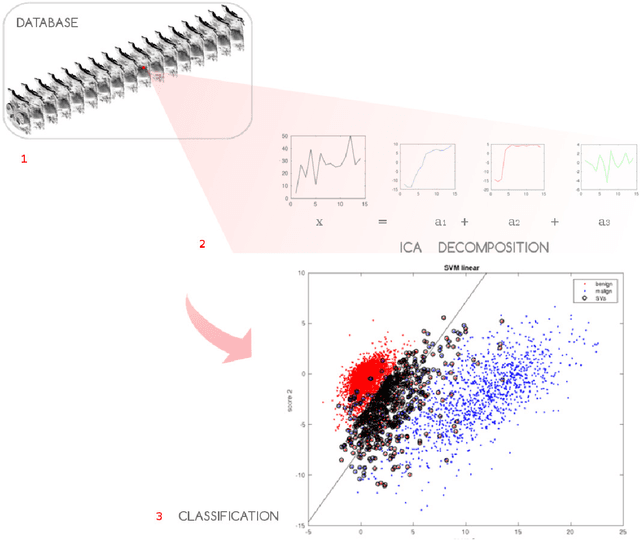
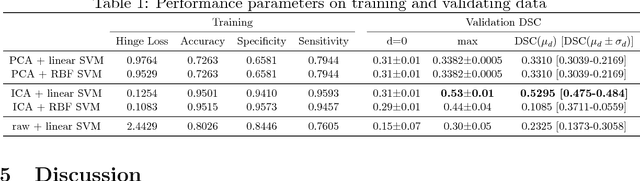
Abstract:Non-mass enhancing lesions (NME) constitute a diagnostic challenge in dynamic contrast enhanced magnetic resonance imaging (DCE-MRI) of the breast. Computer Aided Diagnosis (CAD) systems provide physicians with advanced tools for analysis, assessment and evaluation that have a significant impact on the diagnostic performance. Here, we propose a new approach to address the challenge of NME detection and segmentation, taking advantage of independent component analysis (ICA) to extract data-driven dynamic lesion characterizations. A set of independent sources was obtained from DCE-MRI dataset of breast patients, and the dynamic behavior of the different tissues was described by multiple dynamic curves, together with a set of eigenimages describing the scores for each voxel. A new test image is projected onto the independent source space using the unmixing matrix, and each voxel is classified by a support vector machine (SVM) that has already been trained with manually delineated data. A solution to the high false positive rate problem is proposed by controlling the SVM hyperplane location, outperforming previously published approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge