Jiuwen Zhu
Aggregative Self-Supervised Feature Learning
Dec 15, 2020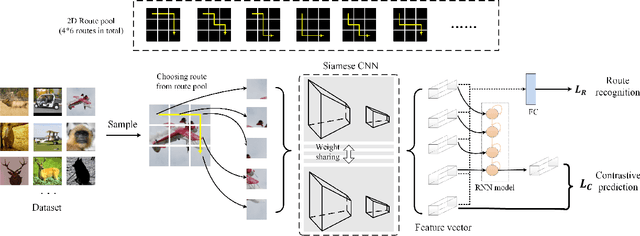
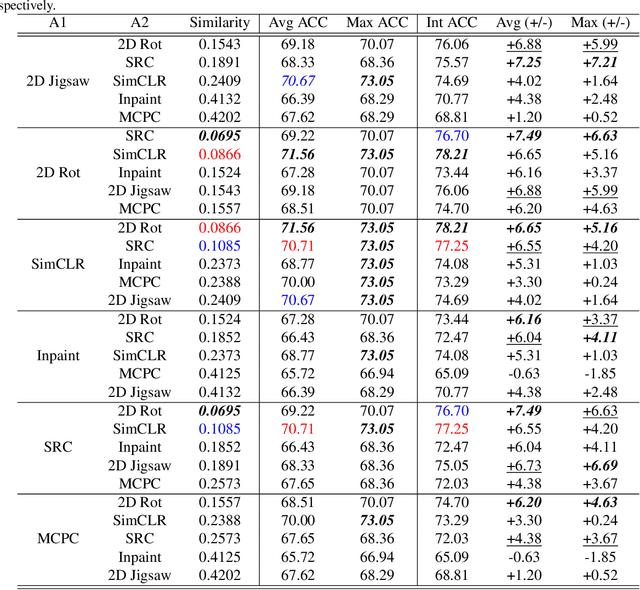
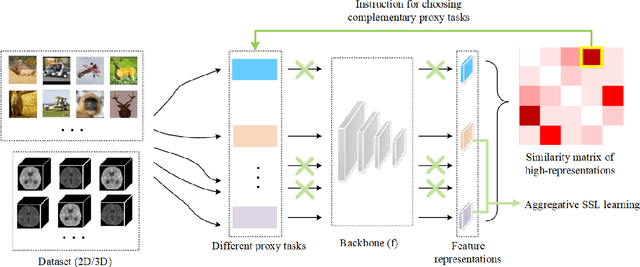
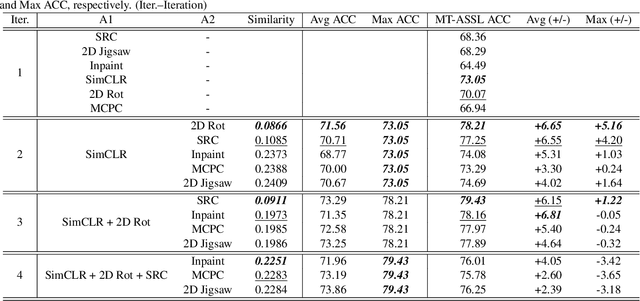
Abstract:Self-supervised learning (SSL) is an efficient approach that addresses the issue of annotation shortage. The key part in SSL is its proxy task that defines the supervisory signals and drives the learning toward effective feature representations. However, most SSL approaches usually focus on a single proxy task, which greatly limits the expressive power of the learned features and therefore deteriorates the network generalization capacity. In this regard, we hereby propose three strategies of aggregation in terms of complementarity of various forms to boost the robustness of self-supervised learned features. In spatial context aggregative SSL, we contribute a heuristic SSL method that integrates two ad-hoc proxy tasks with spatial context complementarity, modeling global and local contextual features, respectively. We then propose a principled framework of multi-task aggregative self-supervised learning to form a unified representation, with an intent of exploiting feature complementarity among different tasks. Finally, in self-aggregative SSL, we propose to self-complement an existing proxy task with an auxiliary loss function based on a linear centered kernel alignment metric, which explicitly promotes the exploring of where are uncovered by the features learned from a proxy task at hand to further boost the modeling capability. Our extensive experiments on 2D natural image and 3D medical image classification tasks under limited annotation scenarios confirm that the proposed aggregation strategies successfully boost the classification accuracy.
Embedding Task Knowledge into 3D Neural Networks via Self-supervised Learning
Jun 10, 2020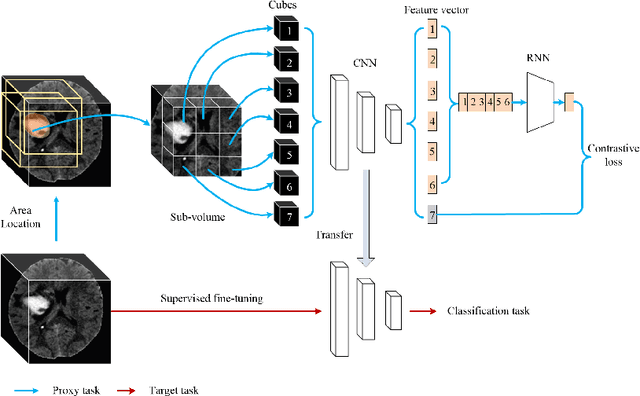
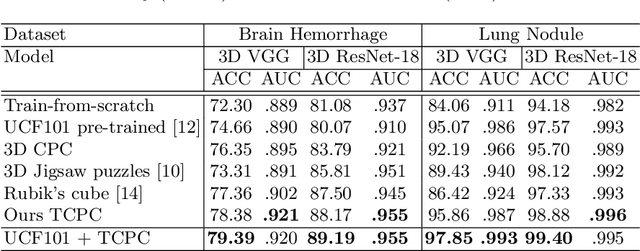
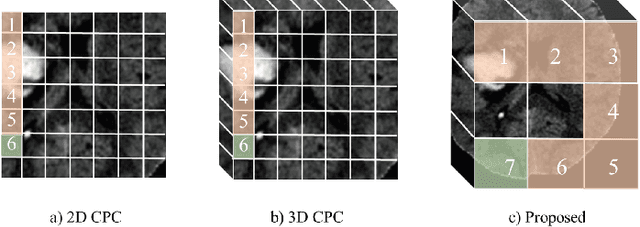
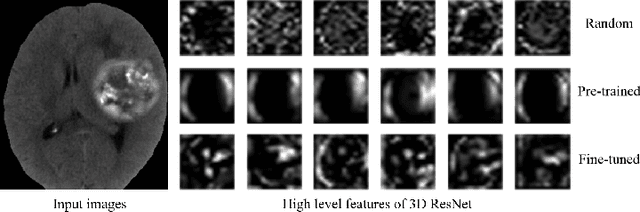
Abstract:Deep learning highly relies on the amount of annotated data. However, annotating medical images is extremely laborious and expensive. To this end, self-supervised learning (SSL), as a potential solution for deficient annotated data, attracts increasing attentions from the community. However, SSL approaches often design a proxy task that is not necessarily related to target task. In this paper, we propose a novel SSL approach for 3D medical image classification, namely Task-related Contrastive Prediction Coding (TCPC), which embeds task knowledge into training 3D neural networks. The proposed TCPC first locates the initial candidate lesions via supervoxel estimation using simple linear iterative clustering. Then, we extract features from the sub-volume cropped around potential lesion areas, and construct a calibrated contrastive predictive coding scheme for self-supervised learning. Extensive experiments are conducted on public and private datasets. The experimental results demonstrate the effectiveness of embedding lesion-related prior-knowledge into neural networks for 3D medical image classification.
Human Recognition Using Face in Computed Tomography
May 28, 2020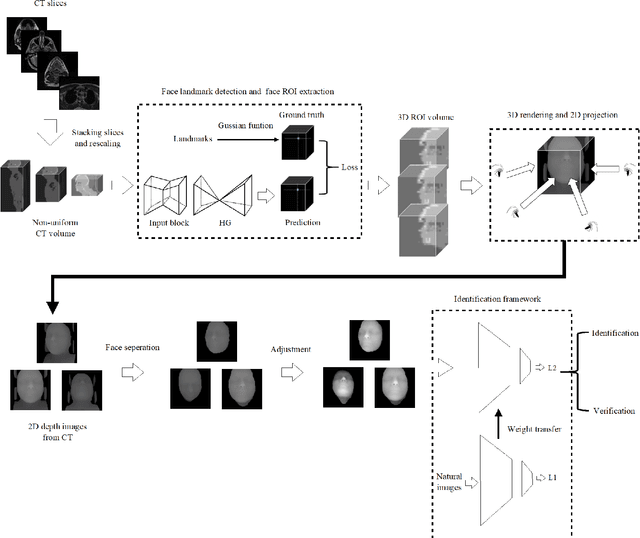
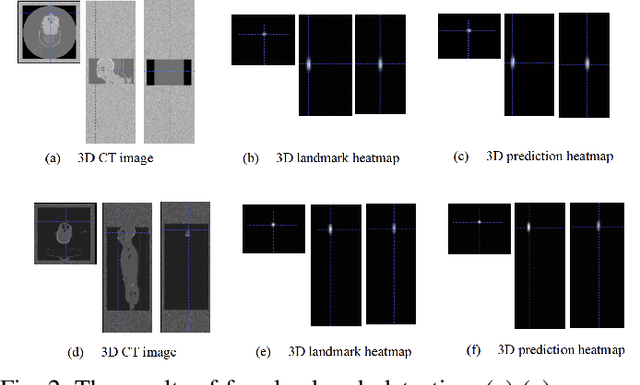
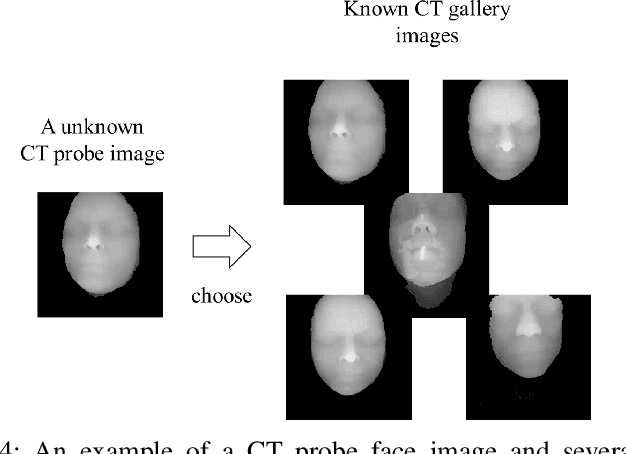
Abstract:With the mushrooming use of computed tomography (CT) images in clinical decision making, management of CT data becomes increasingly difficult. From the patient identification perspective, using the standard DICOM tag to track patient information is challenged by issues such as misspelling, lost file, site variation, etc. In this paper, we explore the feasibility of leveraging the faces in 3D CT images as biometric features. Specifically, we propose an automatic processing pipeline that first detects facial landmarks in 3D for ROI extraction and then generates aligned 2D depth images, which are used for automatic recognition. To boost the recognition performance, we employ transfer learning to reduce the data sparsity issue and to introduce a group sampling strategy to increase inter-class discrimination when training the recognition network. Our proposed method is capable of capturing underlying identity characteristics in medical images while reducing memory consumption. To test its effectiveness, we curate 600 3D CT images of 280 patients from multiple sources for performance evaluation. Experimental results demonstrate that our method achieves a 1:56 identification accuracy of 92.53% and a 1:1 verification accuracy of 96.12%, outperforming other competing approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge