Jessica M. Sin
A Benchmark for Long-Form Medical Question Answering
Nov 14, 2024



Abstract:There is a lack of benchmarks for evaluating large language models (LLMs) in long-form medical question answering (QA). Most existing medical QA evaluation benchmarks focus on automatic metrics and multiple-choice questions. While valuable, these benchmarks fail to fully capture or assess the complexities of real-world clinical applications where LLMs are being deployed. Furthermore, existing studies on evaluating long-form answer generation in medical QA are primarily closed-source, lacking access to human medical expert annotations, which makes it difficult to reproduce results and enhance existing baselines. In this work, we introduce a new publicly available benchmark featuring real-world consumer medical questions with long-form answer evaluations annotated by medical doctors. We performed pairwise comparisons of responses from various open and closed-source medical and general-purpose LLMs based on criteria such as correctness, helpfulness, harmfulness, and bias. Additionally, we performed a comprehensive LLM-as-a-judge analysis to study the alignment between human judgments and LLMs. Our preliminary results highlight the strong potential of open LLMs in medical QA compared to leading closed models. Code & Data: https://github.com/lavita-ai/medical-eval-sphere
Sensitivity and Specificity Evaluation of Deep Learning Models for Detection of Pneumoperitoneum on Chest Radiographs
Oct 17, 2020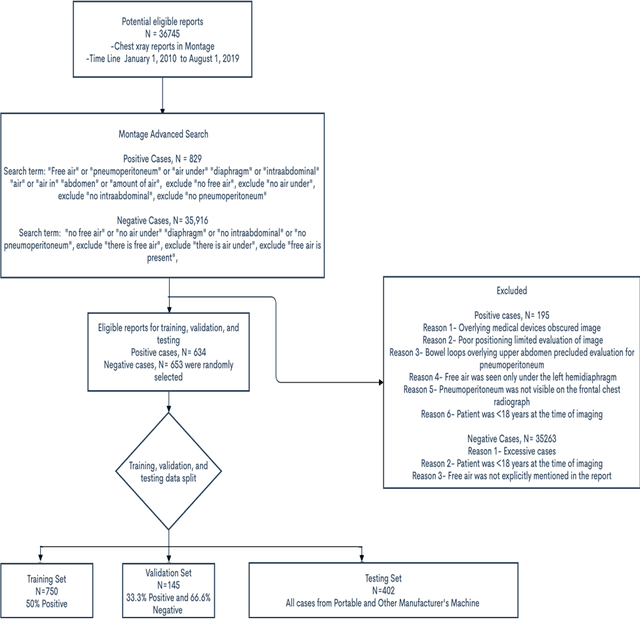
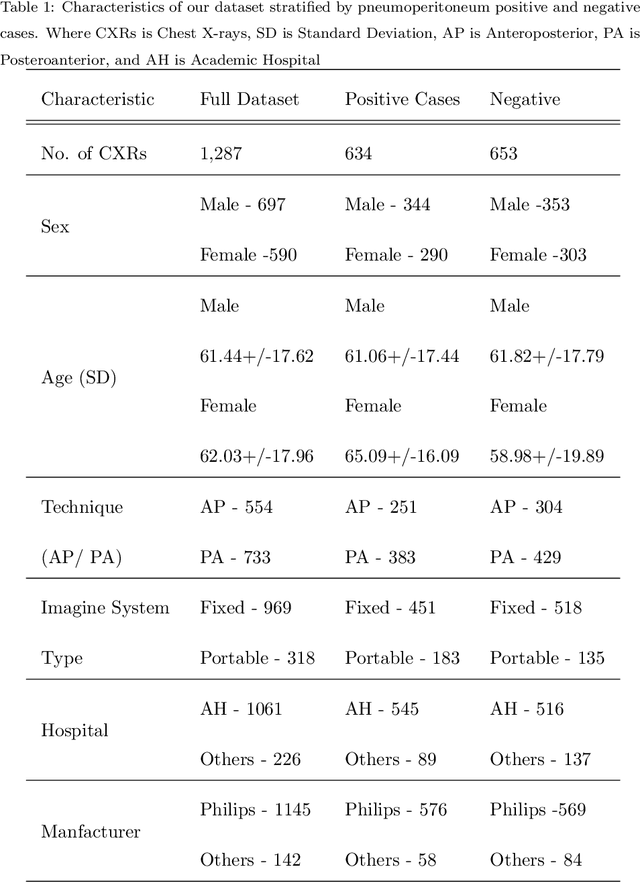
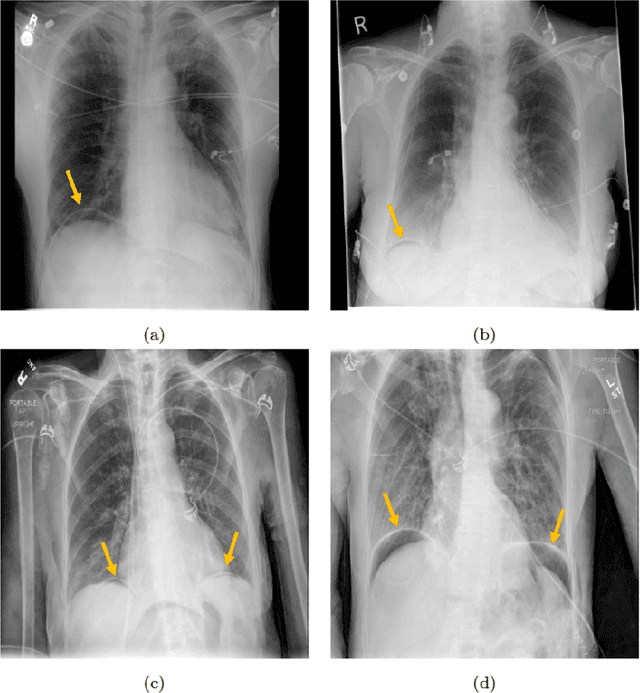
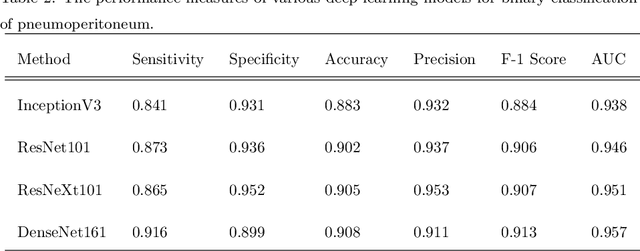
Abstract:Background: Deep learning has great potential to assist with detecting and triaging critical findings such as pneumoperitoneum on medical images. To be clinically useful, the performance of this technology still needs to be validated for generalizability across different types of imaging systems. Materials and Methods: This retrospective study included 1,287 chest X-ray images of patients who underwent initial chest radiography at 13 different hospitals between 2011 and 2019. The chest X-ray images were labelled independently by four radiologist experts as positive or negative for pneumoperitoneum. State-of-the-art deep learning models (ResNet101, InceptionV3, DenseNet161, and ResNeXt101) were trained on a subset of this dataset, and the automated classification performance was evaluated on the rest of the dataset by measuring the AUC, sensitivity, and specificity for each model. Furthermore, the generalizability of these deep learning models was assessed by stratifying the test dataset according to the type of the utilized imaging systems. Results: All deep learning models performed well for identifying radiographs with pneumoperitoneum, while DenseNet161 achieved the highest AUC of 95.7%, Specificity of 89.9%, and Sensitivity of 91.6%. DenseNet161 model was able to accurately classify radiographs from different imaging systems (Accuracy: 90.8%), while it was trained on images captured from a specific imaging system from a single institution. This result suggests the generalizability of our model for learning salient features in chest X-ray images to detect pneumoperitoneum, independent of the imaging system.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge