Jesper H. Pedersen
Extraction of Airways using Graph Neural Networks
Apr 12, 2018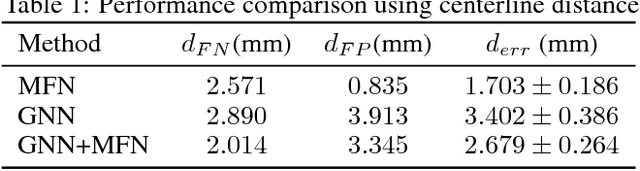
Abstract:We present extraction of tree structures, such as airways, from image data as a graph refinement task. To this end, we propose a graph auto-encoder model that uses an encoder based on graph neural networks (GNNs) to learn embeddings from input node features and a decoder to predict connections between nodes. Performance of the GNN model is compared with mean-field networks in their ability to extract airways from 3D chest CT scans.
Mean Field Network based Graph Refinement with application to Airway Tree Extraction
Apr 10, 2018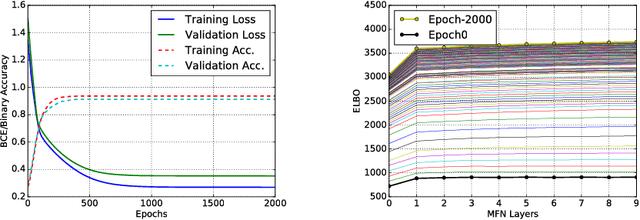

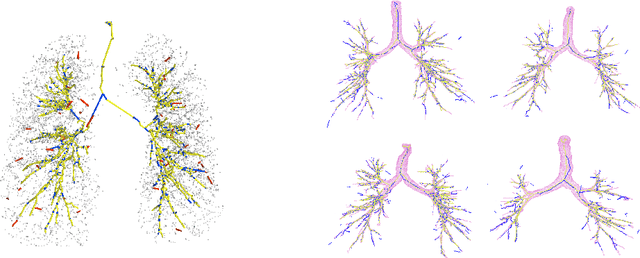
Abstract:We present tree extraction in 3D images as a graph refinement task, of obtaining a subgraph from an over-complete input graph. To this end, we formulate an approximate Bayesian inference framework on undirected graphs using mean field approximation (MFA). Mean field networks are used for inference based on the interpretation that iterations of MFA can be seen as feed-forward operations in a neural network. This allows us to learn the model parameters from training data using back-propagation algorithm. We demonstrate usefulness of the model to extract airway trees from 3D chest CT data. We first obtain probability images using a voxel classifier that distinguishes airways from background and use Bayesian smoothing to model individual airway branches. This yields us joint Gaussian density estimates of position, orientation and scale as node features of the input graph. Performance of the method is compared with two methods: the first uses probability images from a trained voxel classifier with region growing, which is similar to one of the best performing methods at EXACT'09 airway challenge, and the second method is based on Bayesian smoothing on these probability images. Using centerline distance as error measure the presented method shows significant improvement compared to these two methods.
Extraction of Airways with Probabilistic State-space Models and Bayesian Smoothing
Aug 07, 2017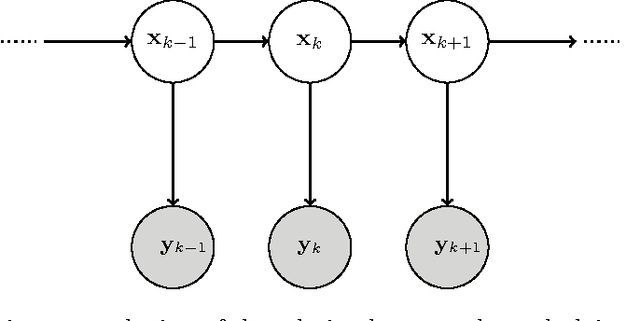
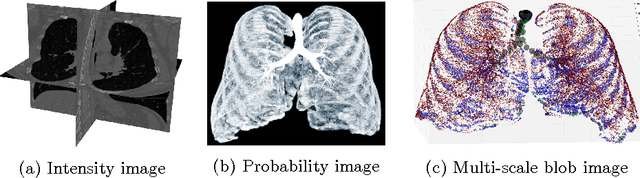
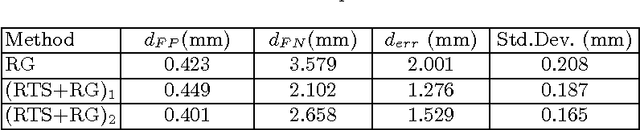
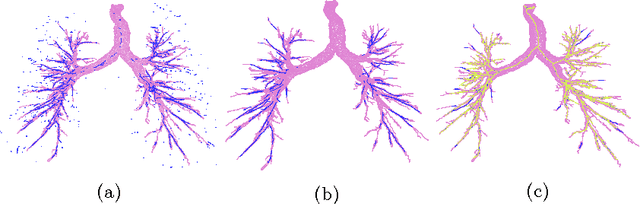
Abstract:Segmenting tree structures is common in several image processing applications. In medical image analysis, reliable segmentations of airways, vessels, neurons and other tree structures can enable important clinical applications. We present a framework for tracking tree structures comprising of elongated branches using probabilistic state-space models and Bayesian smoothing. Unlike most existing methods that proceed with sequential tracking of branches, we present an exploratory method, that is less sensitive to local anomalies in the data due to acquisition noise and/or interfering structures. The evolution of individual branches is modelled using a process model and the observed data is incorporated into the update step of the Bayesian smoother using a measurement model that is based on a multi-scale blob detector. Bayesian smoothing is performed using the RTS (Rauch-Tung-Striebel) smoother, which provides Gaussian density estimates of branch states at each tracking step. We select likely branch seed points automatically based on the response of the blob detection and track from all such seed points using the RTS smoother. We use covariance of the marginal posterior density estimated for each branch to discriminate false positive and true positive branches. The method is evaluated on 3D chest CT scans to track airways. We show that the presented method results in additional branches compared to a baseline method based on region growing on probability images.
Extraction of airway trees using multiple hypothesis tracking and template matching
Nov 24, 2016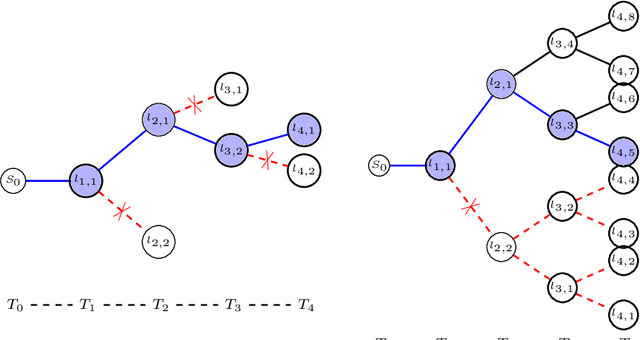
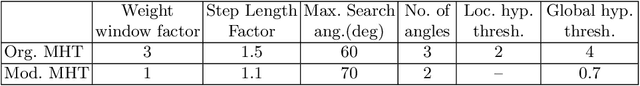
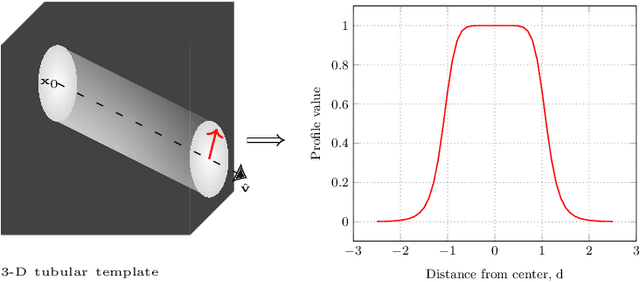
Abstract:Knowledge of airway tree morphology has important clinical applications in diagnosis of chronic obstructive pulmonary disease. We present an automatic tree extraction method based on multiple hypothesis tracking and template matching for this purpose and evaluate its performance on chest CT images. The method is adapted from a semi-automatic method devised for vessel segmentation. Idealized tubular templates are constructed that match airway probability obtained from a trained classifier and ranked based on their relative significance. Several such regularly spaced templates form the local hypotheses used in constructing a multiple hypothesis tree, which is then traversed to reach decisions. The proposed modifications remove the need for local thresholding of hypotheses as decisions are made entirely based on statistical comparisons involving the hypothesis tree. The results show improvements in performance when compared to the original method and region growing on intensity images. We also compare the method with region growing on the probability images, where the presented method does not show substantial improvement, but we expect it to be less sensitive to local anomalies in the data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge