James Green
Sleep Stage Classification using Multimodal Embedding Fusion from EOG and PSM
Jun 07, 2025Abstract:Accurate sleep stage classification is essential for diagnosing sleep disorders, particularly in aging populations. While traditional polysomnography (PSG) relies on electroencephalography (EEG) as the gold standard, its complexity and need for specialized equipment make home-based sleep monitoring challenging. To address this limitation, we investigate the use of electrooculography (EOG) and pressure-sensitive mats (PSM) as less obtrusive alternatives for five-stage sleep-wake classification. This study introduces a novel approach that leverages ImageBind, a multimodal embedding deep learning model, to integrate PSM data with dual-channel EOG signals for sleep stage classification. Our method is the first reported approach that fuses PSM and EOG data for sleep stage classification with ImageBind. Our results demonstrate that fine-tuning ImageBind significantly improves classification accuracy, outperforming existing models based on single-channel EOG (DeepSleepNet), exclusively PSM data (ViViT), and other multimodal deep learning approaches (MBT). Notably, the model also achieved strong performance without fine-tuning, highlighting its adaptability to specific tasks with limited labeled data, making it particularly advantageous for medical applications. We evaluated our method using 85 nights of patient recordings from a sleep clinic. Our findings suggest that pre-trained multimodal embedding models, even those originally developed for non-medical domains, can be effectively adapted for sleep staging, with accuracies approaching systems that require complex EEG data.
Sleep Position Classification using Transfer Learning for Bed-based Pressure Sensors
May 12, 2025Abstract:Bed-based pressure-sensitive mats (PSMs) offer a non-intrusive way of monitoring patients during sleep. We focus on four-way sleep position classification using data collected from a PSM placed under a mattress in a sleep clinic. Sleep positions can affect sleep quality and the prevalence of sleep disorders, such as apnea. Measurements were performed on patients with suspected sleep disorders referred for assessments at a sleep clinic. Training deep learning models can be challenging in clinical settings due to the need for large amounts of labeled data. To overcome the shortage of labeled training data, we utilize transfer learning to adapt pre-trained deep learning models to accurately estimate sleep positions from a low-resolution PSM dataset collected in a polysomnography sleep lab. Our approach leverages Vision Transformer models pre-trained on ImageNet using masked autoencoding (ViTMAE) and a pre-trained model for human pose estimation (ViTPose). These approaches outperform previous work from PSM-based sleep pose classification using deep learning (TCN) as well as traditional machine learning models (SVM, XGBoost, Random Forest) that use engineered features. We evaluate the performance of sleep position classification from 112 nights of patient recordings and validate it on a higher resolution 13-patient dataset. Despite the challenges of differentiating between sleep positions from low-resolution PSM data, our approach shows promise for real-world deployment in clinical settings
Heter-LP: A heterogeneous label propagation algorithm and its application in drug repositioning
Nov 08, 2016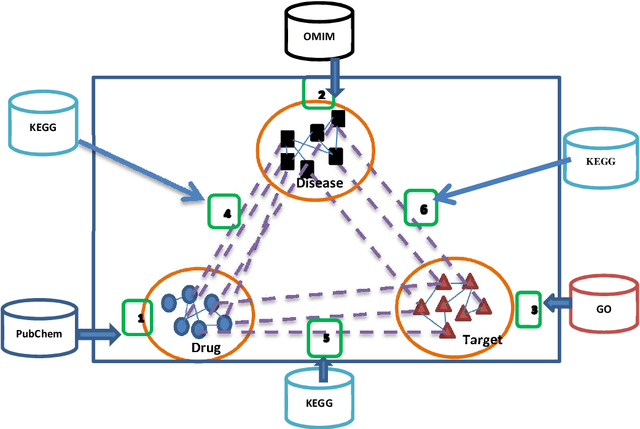
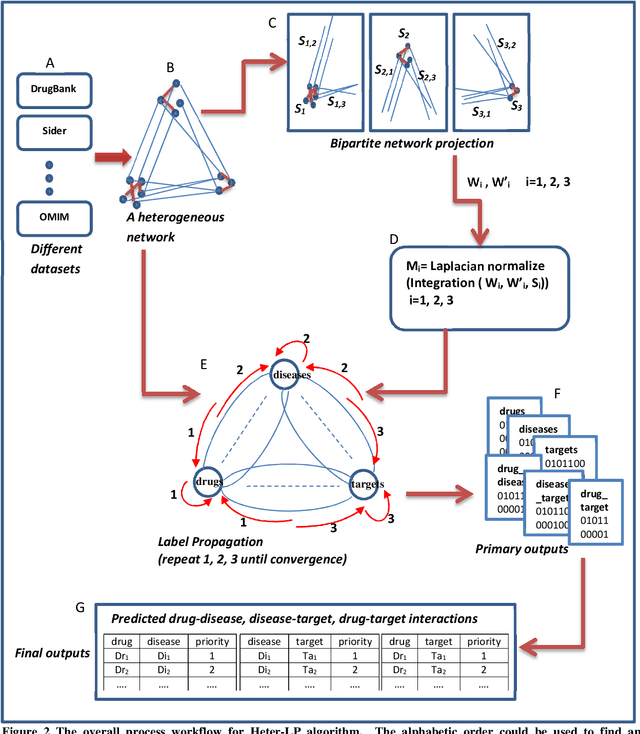
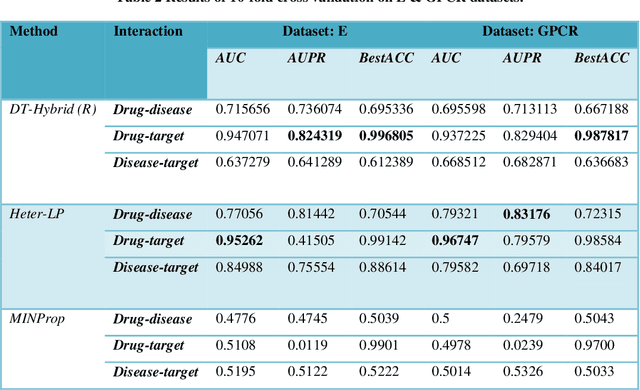
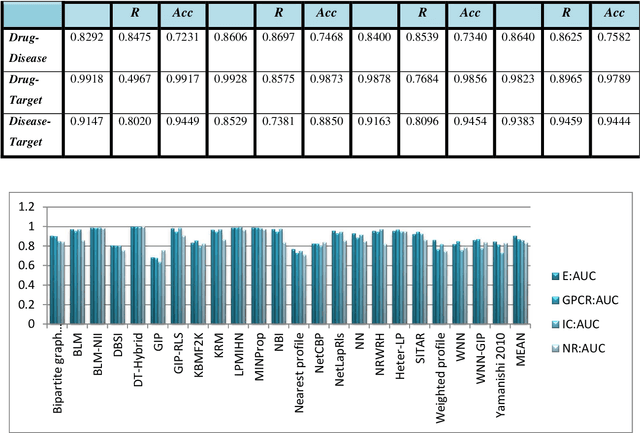
Abstract:Drug repositioning offers an effective solution to drug discovery, saving both time and resources by finding new indications for existing drugs. Typically, a drug takes effect via its protein targets in the cell. As a result, it is necessary for drug development studies to conduct an investigation into the interrelationships of drugs, protein targets, and diseases. Although previous studies have made a strong case for the effectiveness of integrative network-based methods for predicting these interrelationships, little progress has been achieved in this regard within drug repositioning research. Moreover, the interactions of new drugs and targets (lacking any known targets and drugs, respectively) cannot be accurately predicted by most established methods. In this paper, we propose a novel semi-supervised heterogeneous label propagation algorithm named Heter-LP, which applies both local as well as global network features for data integration. To predict drug-target, disease-target, and drug-disease associations, we use information about drugs, diseases, and targets as collected from multiple sources at different levels. Our algorithm integrates these various types of data into a heterogeneous network and implements a label propagation algorithm to find new interactions. Statistical analyses of 10-fold cross-validation results and experimental analysis support the effectiveness of the proposed algorithm.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge