Iris Wang
OscNet v1.5: Energy Efficient Hopfield Network on CMOS Oscillators for Image Classification
Jun 14, 2025Abstract:Machine learning has achieved remarkable advancements but at the cost of significant computational resources. This has created an urgent need for a novel and energy-efficient computational fabric. CMOS Oscillator Networks (OscNet) is a brain inspired and specially designed hardware for low energy consumption. In this paper, we propose a Hopfield Network based machine learning algorithm that can be implemented on OscNet. The network is trained using forward propagation alone to learn sparsely connected weights, yet achieves an 8% improvement in accuracy compared to conventional deep learning models on MNIST dataset. OscNet v1.5 achieves competitive accuracy on MNIST and is well-suited for implementation using CMOS-compatible ring oscillator arrays with SHIL. In oscillator-based implementation, we utilize only 24% of the connections used in a fully connected Hopfield network, with merely a 0.1% drop in accuracy. OscNet v1.5 relies solely on forward propagation and employs sparse connections, making it an energy-efficient machine learning pipeline designed for CMOS oscillator computing. The repository for OscNet family is: https://github.com/RussRobin/OscNet.
Application of Deep Learning on Predicting Prognosis of Acute Myeloid Leukemia with Cytogenetics, Age, and Mutations
Oct 30, 2018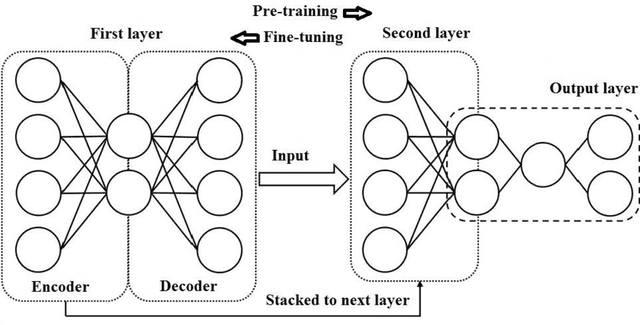
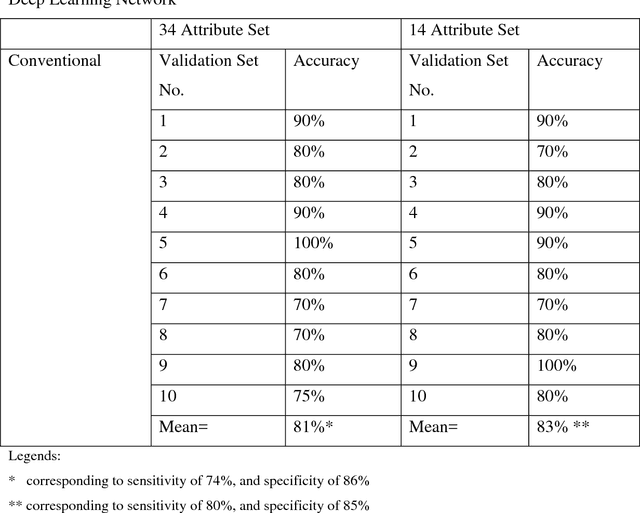
Abstract:We explore how Deep Learning (DL) can be utilized to predict prognosis of acute myeloid leukemia (AML). Out of TCGA (The Cancer Genome Atlas) database, 94 AML cases are used in this study. Input data include age, 10 common cytogenetic and 23 most common mutation results; output is the prognosis (diagnosis to death, DTD). In our DL network, autoencoders are stacked to form a hierarchical DL model from which raw data are compressed and organized and high-level features are extracted. The network is written in R language and is designed to predict prognosis of AML for a given case (DTD of more than or less than 730 days). The DL network achieves an excellent accuracy of 83% in predicting prognosis. As a proof-of-concept study, our preliminary results demonstrate a practical application of DL in future practice of prognostic prediction using next-gen sequencing (NGS) data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge