Hubert Cochet
EPIONE
Constraint-Based Model in Multimodal Learning to Improve Ventricular Arrhythmia Prediction
Dec 18, 2024Abstract:Cardiac disease evaluation depends on multiple diagnostic modalities: electrocardiogram (ECG) to diagnose abnormal heart rhythms, and imaging modalities such as Magnetic Resonance Imaging (MRI), Computed Tomography (CT) and echocardiography to detect signs of structural abnormalities. Each of these modalities brings complementary information for a better diagnosis of cardiac dysfunction. However, training a machine learning (ML) model with data from multiple modalities is a challenging task, as it increases the dimension space, while keeping constant the number of samples. In fact, as the dimension of the input space increases, the volume of data required for accurate generalisation grows exponentially. In this work, we address this issue, for the application of Ventricular Arrhythmia (VA) prediction, based on the combined clinical and CT imaging features, where we constrained the learning process on medical images (CT) based on the prior knowledge acquired from clinical data. The VA classifier is fed with features extracted from a 3D myocardium thickness map (TM) of the left ventricle. The TM is generated by our pipeline from the imaging input and a Graph Convolutional Network is used as the feature extractor of the 3D TM. We introduce a novel Sequential Fusion method and evaluate its performance against traditional Early Fusion techniques and single-modality models. The crossvalidation results show that the Sequential Fusion model achieved the highest average scores of 80.7% $\pm$ 4.4 Sensitivity and 73.1% $\pm$ 6.0 F1 score, outperforming the Early Fusion model at 65.0% $\pm$ 8.9 Sensitivity and 63.1% $\pm$ 6.3 F1 score. Both fusion models achieved better scores than the single-modality models, where the average Sensitivity and F1 score are 62.8% $\pm$ 10.1; 52.1% $\pm$ 6.5 for the clinical data modality and 62.9% $\pm$ 6.3; 60.7% $\pm$ 5.3 for the medical images modality.
Automatically Segmenting the Left Atrium from Cardiac Images Using Successive 3D U-Nets and a Contour Loss
Dec 06, 2018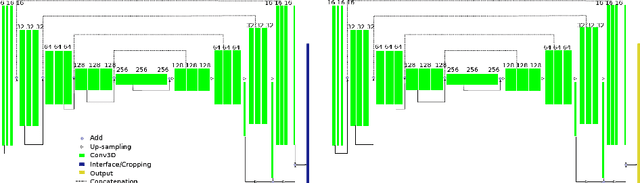
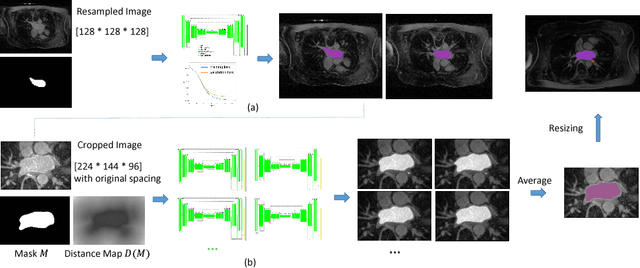
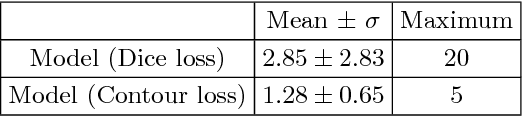
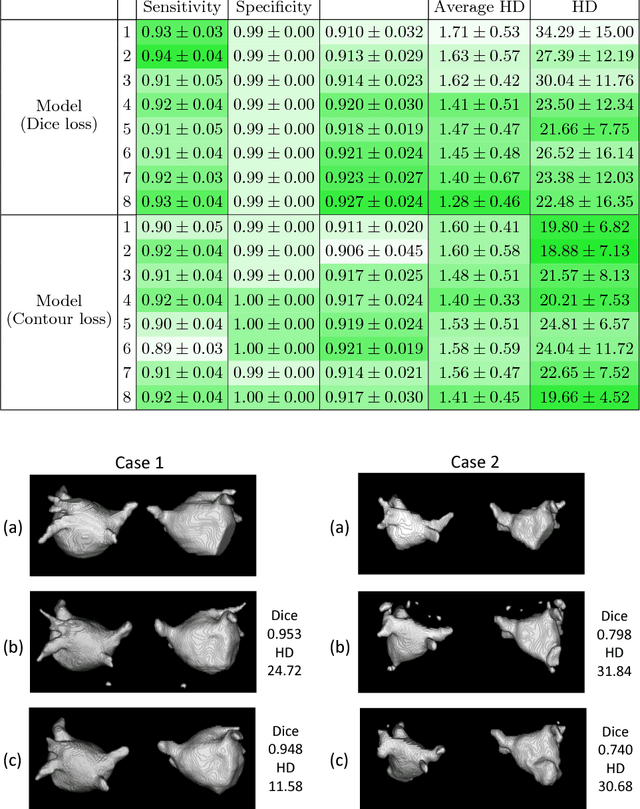
Abstract:Radiological imaging offers effective measurement of anatomy, which is useful in disease diagnosis and assessment. Previous study has shown that the left atrial wall remodeling can provide information to predict treatment outcome in atrial fibrillation. Nevertheless, the segmentation of the left atrial structures from medical images is still very time-consuming. Current advances in neural network may help creating automatic segmentation models that reduce the workload for clinicians. In this preliminary study, we propose automated, two-stage, three-dimensional U-Nets with convolutional neural network, for the challenging task of left atrial segmentation. Unlike previous two-dimensional image segmentation methods, we use 3D U-Nets to obtain the heart cavity directly in 3D. The dual 3D U-Net structure consists of, a first U-Net to coarsely segment and locate the left atrium, and a second U-Net to accurately segment the left atrium under higher resolution. In addition, we introduce a Contour loss based on additional distance information to adjust the final segmentation. We randomly split the data into training datasets (80 subjects) and validation datasets (20 subjects) to train multiple models, with different augmentation setting. Experiments show that the average Dice coefficients for validation datasets are around 0.91 - 0.92, the sensitivity around 0.90-0.94 and the specificity 0.99. Compared with traditional Dice loss, models trained with Contour loss in general offer smaller Hausdorff distance with similar Dice coefficient, and have less connected components in predictions. Finally, we integrate several trained models in an ensemble prediction to segment testing datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge