Hossein Estiri
A Hybrid Enumeration Framework for Optimal Counterfactual Generation in Post-Acute COVID-19 Heart Failure
Oct 21, 2025Abstract:Counterfactual inference provides a mathematical framework for reasoning about hypothetical outcomes under alternative interventions, bridging causal reasoning and predictive modeling. We present a counterfactual inference framework for individualized risk estimation and intervention analysis, illustrated through a clinical application to post-acute sequelae of COVID-19 (PASC) among patients with pre-existing heart failure (HF). Using longitudinal diagnosis, laboratory, and medication data from a large health-system cohort, we integrate regularized predictive modeling with counterfactual search to identify actionable pathways to PASC-related HF hospital admissions. The framework combines exact enumeration with optimization-based methods, including the Nearest Instance Counterfactual Explanations (NICE) and Multi-Objective Counterfactuals (MOC) algorithms, to efficiently explore high-dimensional intervention spaces. Applied to more than 2700 individuals with confirmed SARS-CoV-2 infection and prior HF, the model achieved strong discriminative performance (AUROC: 0.88, 95% CI: 0.84-0.91) and generated interpretable, patient-specific counterfactuals that quantify how modifying comorbidity patterns or treatment factors could alter predicted outcomes. This work demonstrates how counterfactual reasoning can be formalized as an optimization problem over predictive functions, offering a rigorous, interpretable, and computationally efficient approach to personalized inference in complex biomedical systems.
The Average Patient Fallacy
Sep 30, 2025
Abstract:Machine learning in medicine is typically optimized for population averages. This frequency weighted training privileges common presentations and marginalizes rare yet clinically critical cases, a bias we call the average patient fallacy. In mixture models, gradients from rare cases are suppressed by prevalence, creating a direct conflict with precision medicine. Clinical vignettes in oncology, cardiology, and ophthalmology show how this yields missed rare responders, delayed recognition of atypical emergencies, and underperformance on vision-threatening variants. We propose operational fixes: Rare Case Performance Gap, Rare Case Calibration Error, a prevalence utility definition of rarity, and clinically weighted objectives that surface ethical priorities. Weight selection should follow structured deliberation. AI in medicine must detect exceptional cases because of their significance.
Signal Fidelity Index-Aware Calibration for Dementia Predictions Across Heterogeneous Real-World Data
Sep 10, 2025



Abstract:\textbf{Background:} Machine learning models trained on electronic health records (EHRs) often degrade across healthcare systems due to distributional shift. A fundamental but underexplored factor is diagnostic signal decay: variability in diagnostic quality and consistency across institutions, which affects the reliability of codes used for training and prediction. \textbf{Objective:} To develop a Signal Fidelity Index (SFI) quantifying diagnostic data quality at the patient level in dementia, and to test SFI-aware calibration for improving model performance across heterogeneous datasets without outcome labels. \textbf{Methods:} We built a simulation framework generating 2,500 synthetic datasets, each with 1,000 patients and realistic demographics, encounters, and coding patterns based on dementia risk factors. The SFI was derived from six interpretable components: diagnostic specificity, temporal consistency, entropy, contextual concordance, medication alignment, and trajectory stability. SFI-aware calibration applied a multiplicative adjustment, optimized across 50 simulation batches. \textbf{Results:} At the optimal parameter ($\alpha$ = 2.0), SFI-aware calibration significantly improved all metrics (p $<$ 0.001). Gains ranged from 10.3\% for Balanced Accuracy to 32.5\% for Recall, with notable increases in Precision (31.9\%) and F1-score (26.1\%). Performance approached reference standards, with F1-score and Recall within 1\% and Balanced Accuracy and Detection Rate improved by 52.3\% and 41.1\%, respectively. \textbf{Conclusions:} Diagnostic signal decay is a tractable barrier to model generalization. SFI-aware calibration provides a practical, label-free strategy to enhance prediction across healthcare contexts, particularly for large-scale administrative datasets lacking outcome labels.
An Agentic AI Workflow for Detecting Cognitive Concerns in Real-world Data
Feb 03, 2025Abstract:Early identification of cognitive concerns is critical but often hindered by subtle symptom presentation. This study developed and validated a fully automated, multi-agent AI workflow using LLaMA 3 8B to identify cognitive concerns in 3,338 clinical notes from Mass General Brigham. The agentic workflow, leveraging task-specific agents that dynamically collaborate to extract meaningful insights from clinical notes, was compared to an expert-driven benchmark. Both workflows achieved high classification performance, with F1-scores of 0.90 and 0.91, respectively. The agentic workflow demonstrated improved specificity (1.00) and achieved prompt refinement in fewer iterations. Although both workflows showed reduced performance on validation data, the agentic workflow maintained perfect specificity. These findings highlight the potential of fully automated multi-agent AI workflows to achieve expert-level accuracy with greater efficiency, offering a scalable and cost-effective solution for detecting cognitive concerns in clinical settings.
tSPM+; a high-performance algorithm for mining transitive sequential patterns from clinical data
Sep 08, 2023



Abstract:The increasing availability of large clinical datasets collected from patients can enable new avenues for computational characterization of complex diseases using different analytic algorithms. One of the promising new methods for extracting knowledge from large clinical datasets involves temporal pattern mining integrated with machine learning workflows. However, mining these temporal patterns is a computational intensive task and has memory repercussions. Current algorithms, such as the temporal sequence pattern mining (tSPM) algorithm, are already providing promising outcomes, but still leave room for optimization. In this paper, we present the tSPM+ algorithm, a high-performance implementation of the tSPM algorithm, which adds a new dimension by adding the duration to the temporal patterns. We show that the tSPM+ algorithm provides a speed up to factor 980 and a up to 48 fold improvement in memory consumption. Moreover, we present a docker container with an R-package, We also provide vignettes for an easy integration into already existing machine learning workflows and use the mined temporal sequences to identify Post COVID-19 patients and their symptoms according to the WHO definition.
Individualized Prediction of COVID-19 Adverse outcomes with MLHO
Aug 10, 2020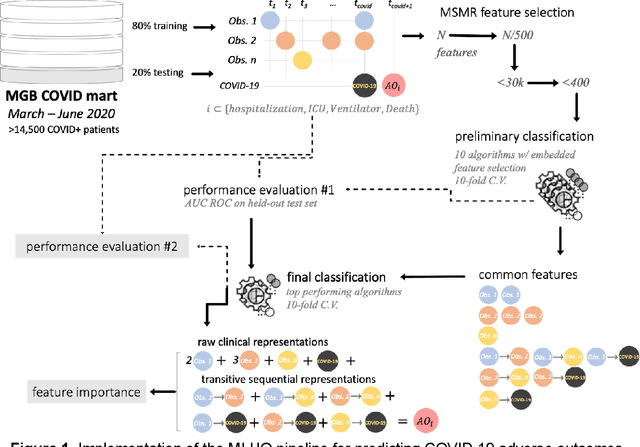
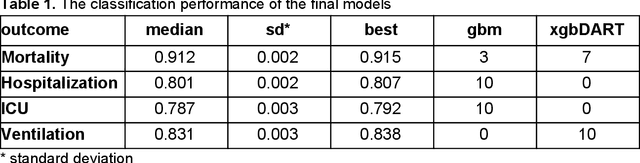
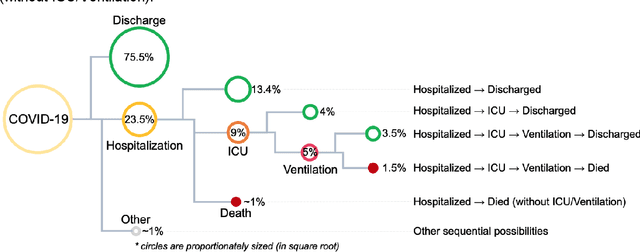
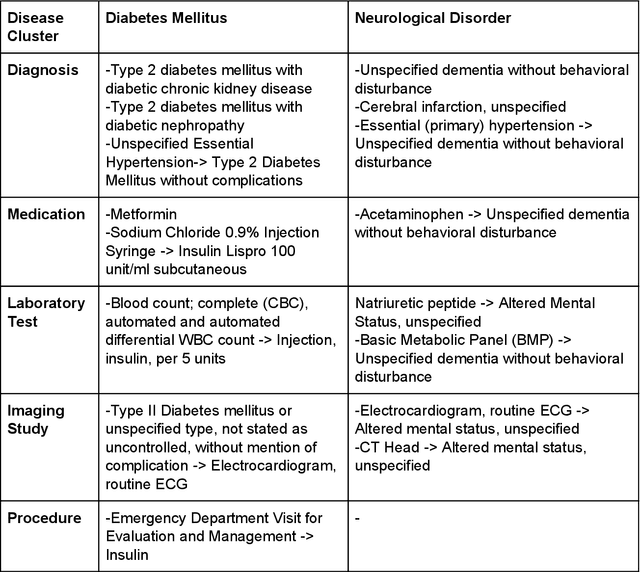
Abstract:The COVID-19 pandemic has devastated the world with health and economic wreckage. Precise estimates of the COVID-19 adverse outcomes on individual patients could have led to better allocation of healthcare resources and more efficient targeted preventive measures. We developed MLHO (pronounced as melo) for predicting patient-level risk of hospitalization, ICU admission, need for mechanical ventilation, and death from patients' past (before COVID-19 infection) medical records. MLHO is an end-to-end Machine Learning pipeline that implements iterative sequential representation mining and feature and model selection to predict health outcomes. MLHO's architecture enables a parallel and outcome-oriented calibration, in which different statistical learning algorithms and vectors of features are simultaneously tested and leveraged to improve prediction of health outcomes. Using clinical data from a large cohort of over 14,000 patients, we modeled the four adverse outcomes utilizing about 600 features representing patients' before-COVID health records. Overall, the best predictions were obtained from extreme and gradient boosting models. The median AUC ROC for mortality prediction was 0.91, while the prediction performance ranged between 0.79 and 0.83 for ICU, hospitalization, and ventilation. We broadly describe the clusters of features that were utilized in modeling and their relative influence on predicting each outcome. As COVID-19 cases are re-surging in the U.S. and around the world, a Machine Learning pipeline like MLHO is crucial to improve our readiness for confronting the potential future waves of COVID-19, as well as other novel infectious diseases that may emerge in the near future.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge