Hamid Kamangir
California Crop Yield Benchmark: Combining Satellite Image, Climate, Evapotranspiration, and Soil Data Layers for County-Level Yield Forecasting of Over 70 Crops
Jun 11, 2025



Abstract:California is a global leader in agricultural production, contributing 12.5% of the United States total output and ranking as the fifth-largest food and cotton supplier in the world. Despite the availability of extensive historical yield data from the USDA National Agricultural Statistics Service, accurate and timely crop yield forecasting remains a challenge due to the complex interplay of environmental, climatic, and soil-related factors. In this study, we introduce a comprehensive crop yield benchmark dataset covering over 70 crops across all California counties from 2008 to 2022. The benchmark integrates diverse data sources, including Landsat satellite imagery, daily climate records, monthly evapotranspiration, and high-resolution soil properties. To effectively learn from these heterogeneous inputs, we develop a multi-modal deep learning model tailored for county-level, crop-specific yield forecasting. The model employs stratified feature extraction and a timeseries encoder to capture spatial and temporal dynamics during the growing season. Static inputs such as soil characteristics and crop identity inform long-term variability. Our approach achieves an overall R2 score of 0.76 across all crops of unseen test dataset, highlighting strong predictive performance across California diverse agricultural regions. This benchmark and modeling framework offer a valuable foundation for advancing agricultural forecasting, climate adaptation, and precision farming. The full dataset and codebase are publicly available at our GitHub repository.
CMAViT: Integrating Climate, Managment, and Remote Sensing Data for Crop Yield Estimation with Multimodel Vision Transformers
Nov 25, 2024



Abstract:Crop yield prediction is essential for agricultural planning but remains challenging due to the complex interactions between weather, climate, and management practices. To address these challenges, we introduce a deep learning-based multi-model called Climate-Management Aware Vision Transformer (CMAViT), designed for pixel-level vineyard yield predictions. CMAViT integrates both spatial and temporal data by leveraging remote sensing imagery and short-term meteorological data, capturing the effects of growing season variations. Additionally, it incorporates management practices, which are represented in text form, using a cross-attention encoder to model their interaction with time-series data. This innovative multi-modal transformer tested on a large dataset from 2016-2019 covering 2,200 hectares and eight grape cultivars including more than 5 million vines, outperforms traditional models like UNet-ConvLSTM, excelling in spatial variability capture and yield prediction, particularly for extreme values in vineyards. CMAViT achieved an R2 of 0.84 and a MAPE of 8.22% on an unseen test dataset. Masking specific modalities lowered performance: excluding management practices, climate data, and both reduced R2 to 0.73, 0.70, and 0.72, respectively, and raised MAPE to 11.92%, 12.66%, and 12.39%, highlighting each modality's importance for accurate yield prediction. Code is available at https://github.com/plant-ai-biophysics-lab/CMAViT.
Simultaneously Predicting Multiple Plant Traits from Multiple Sensors via Deformable CNN Regression
Dec 06, 2021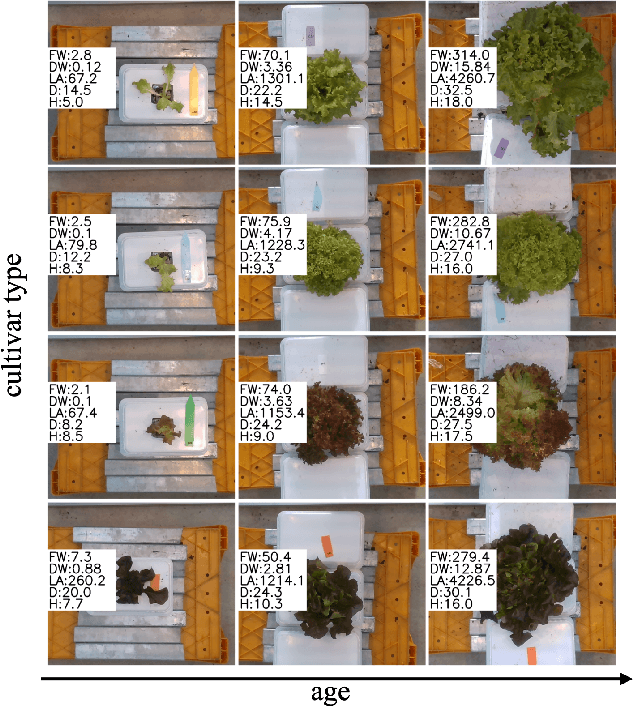
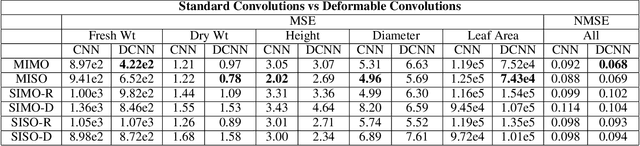
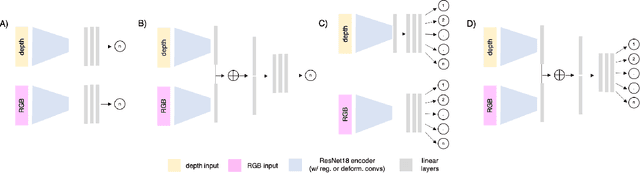
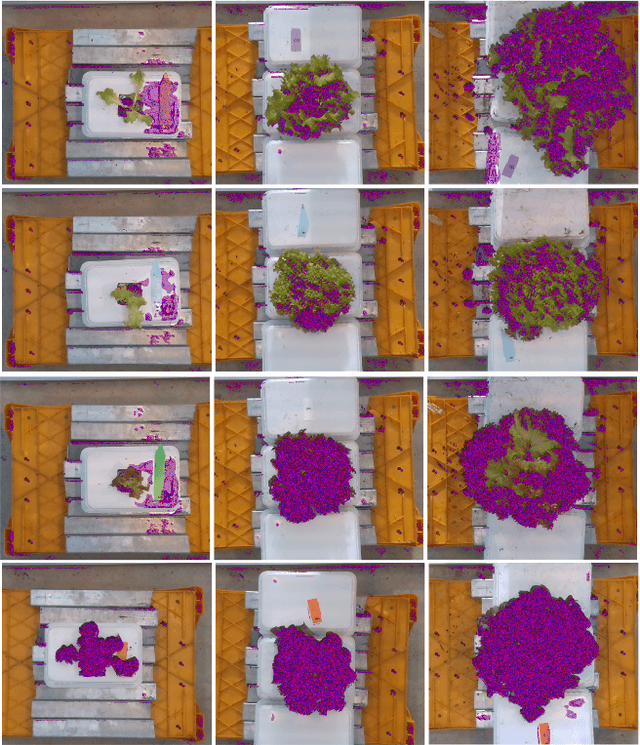
Abstract:Trait measurement is critical for the plant breeding and agricultural production pipeline. Typically, a suite of plant traits is measured using laborious manual measurements and then used to train and/or validate higher throughput trait estimation techniques. Here, we introduce a relatively simple convolutional neural network (CNN) model that accepts multiple sensor inputs and predicts multiple continuous trait outputs - i.e. a multi-input, multi-output CNN (MIMO-CNN). Further, we introduce deformable convolutional layers into this network architecture (MIMO-DCNN) to enable the model to adaptively adjust its receptive field, model complex variable geometric transformations in the data, and fine-tune the continuous trait outputs. We examine how the MIMO-CNN and MIMO-DCNN models perform on a multi-input (i.e. RGB and depth images), multi-trait output lettuce dataset from the 2021 Autonomous Greenhouse Challenge. Ablation studies were conducted to examine the effect of using single versus multiple inputs, and single versus multiple outputs. The MIMO-DCNN model resulted in a normalized mean squared error (NMSE) of 0.068 - a substantial improvement over the top 2021 leaderboard score of 0.081. Open-source code is provided.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge