H. R. Tizhoosh
Kimia Lab, University of Waterloo, Waterloo, ON, Canada, Vector Institute, MaRS Centre, Toronto, Canada
Learning Binary and Sparse Permutation-Invariant Representations for Fast and Memory Efficient Whole Slide Image Search
Aug 29, 2022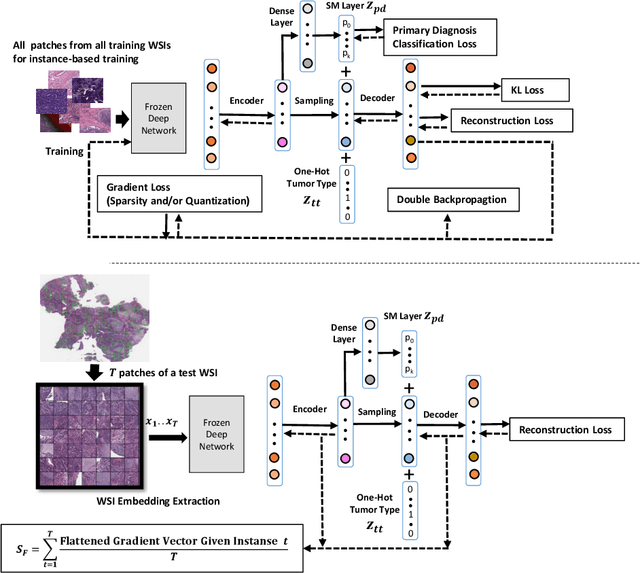
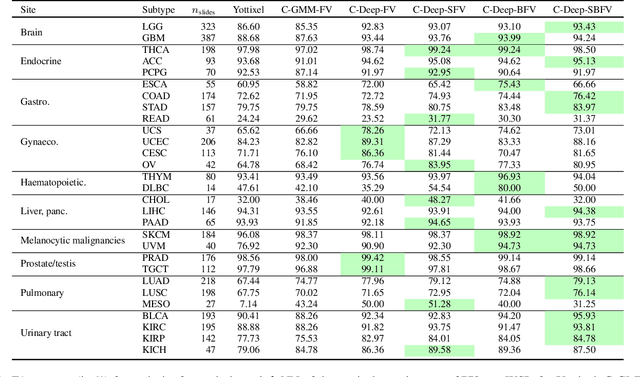
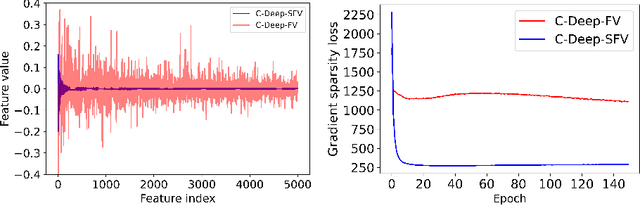
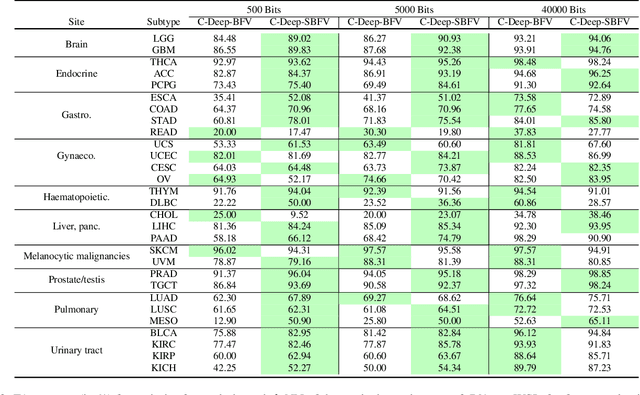
Abstract:Learning suitable Whole slide images (WSIs) representations for efficient retrieval systems is a non-trivial task. The WSI embeddings obtained from current methods are in Euclidean space not ideal for efficient WSI retrieval. Furthermore, most of the current methods require high GPU memory due to the simultaneous processing of multiple sets of patches. To address these challenges, we propose a novel framework for learning binary and sparse WSI representations utilizing a deep generative modelling and the Fisher Vector. We introduce new loss functions for learning sparse and binary permutation-invariant WSI representations that employ instance-based training achieving better memory efficiency. The learned WSI representations are validated on The Cancer Genomic Atlas (TCGA) and Liver-Kidney-Stomach (LKS) datasets. The proposed method outperforms Yottixel (a recent search engine for histopathology images) both in terms of retrieval accuracy and speed. Further, we achieve competitive performance against SOTA on the public benchmark LKS dataset for WSI classification.
Cluster Based Secure Multi-Party Computation in Federated Learning for Histopathology Images
Aug 21, 2022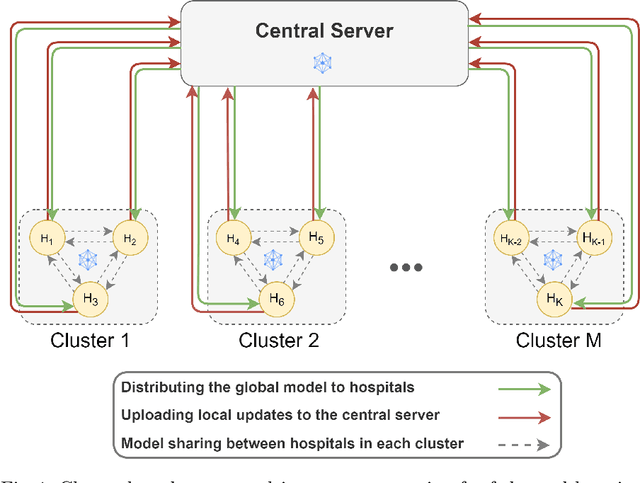
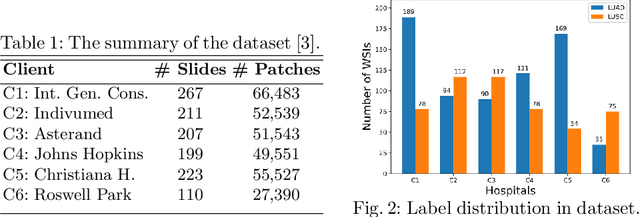
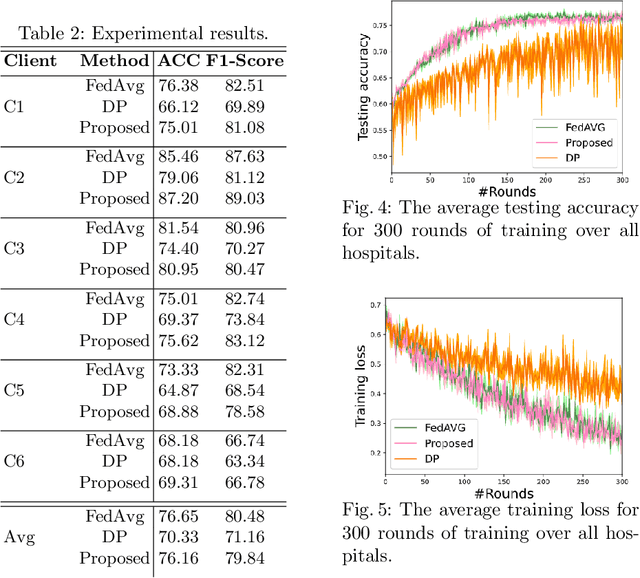
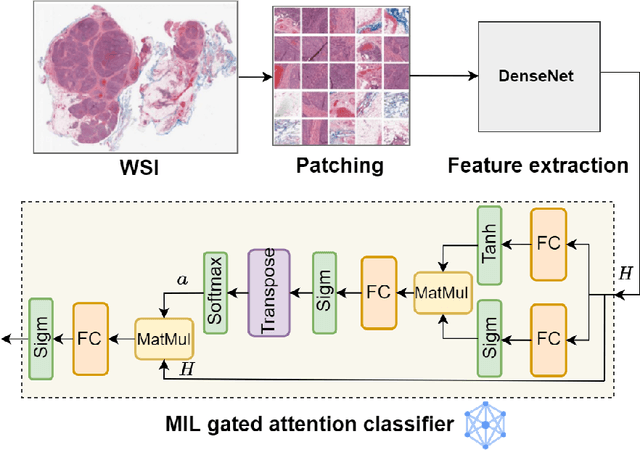
Abstract:Federated learning (FL) is a decentralized method enabling hospitals to collaboratively learn a model without sharing private patient data for training. In FL, participant hospitals periodically exchange training results rather than training samples with a central server. However, having access to model parameters or gradients can expose private training data samples. To address this challenge, we adopt secure multiparty computation (SMC) to establish a privacy-preserving federated learning framework. In our proposed method, the hospitals are divided into clusters. After local training, each hospital splits its model weights among other hospitals in the same cluster such that no single hospital can retrieve other hospitals' weights on its own. Then, all hospitals sum up the received weights, sending the results to the central server. Finally, the central server aggregates the results, retrieving the average of models' weights and updating the model without having access to individual hospitals' weights. We conduct experiments on a publicly available repository, The Cancer Genome Atlas (TCGA). We compare the performance of the proposed framework with differential privacy and federated averaging as the baseline. The results reveal that compared to differential privacy, our framework can achieve higher accuracy with no privacy leakage risk at a cost of higher communication overhead.
Monitoring Shortcut Learning using Mutual Information
Jun 27, 2022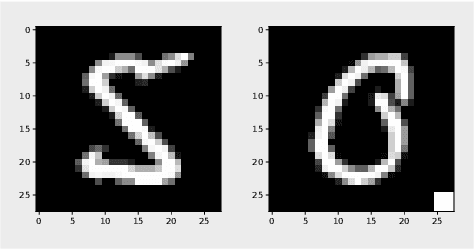
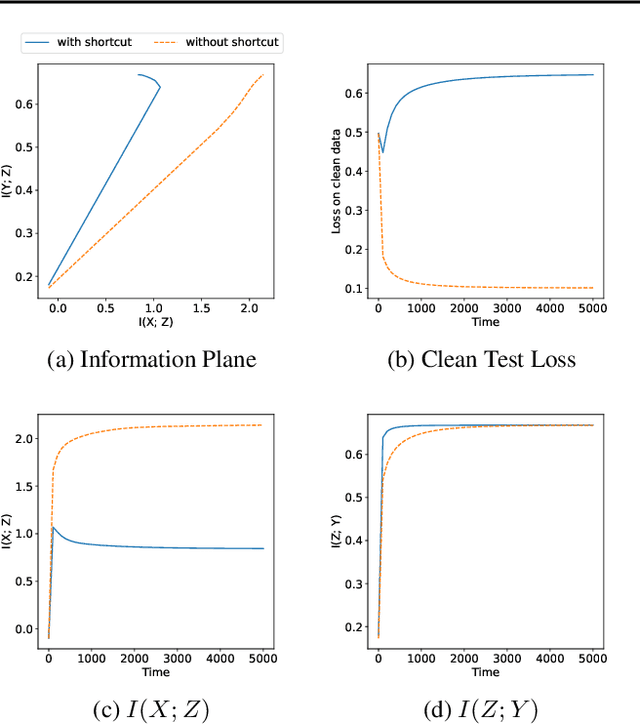
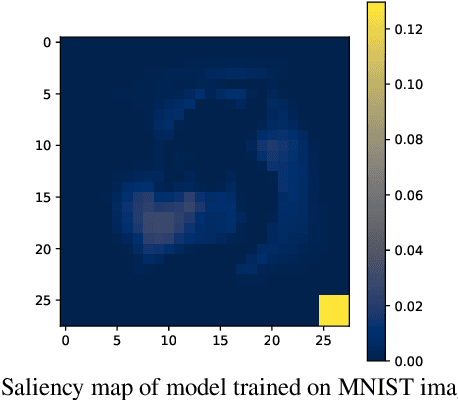
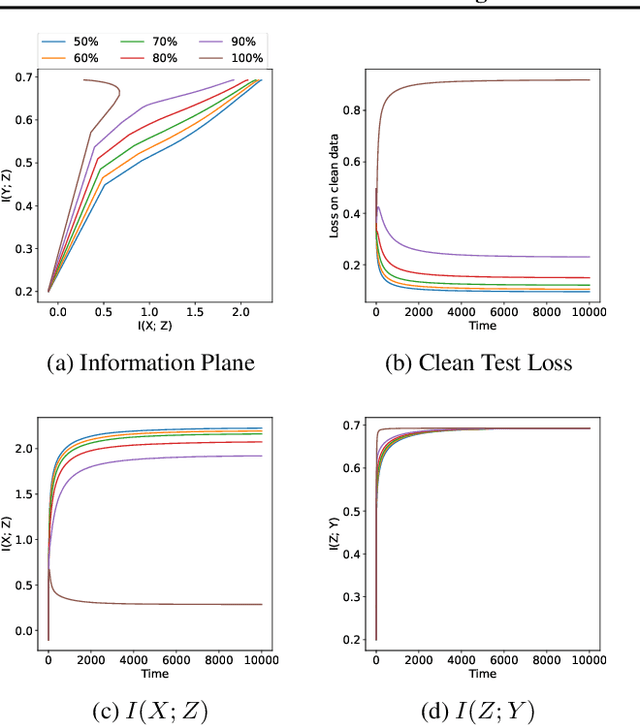
Abstract:The failure of deep neural networks to generalize to out-of-distribution data is a well-known problem and raises concerns about the deployment of trained networks in safety-critical domains such as healthcare, finance and autonomous vehicles. We study a particular kind of distribution shift $\unicode{x2013}$ shortcuts or spurious correlations in the training data. Shortcut learning is often only exposed when models are evaluated on real-world data that does not contain the same spurious correlations, posing a serious dilemma for AI practitioners to properly assess the effectiveness of a trained model for real-world applications. In this work, we propose to use the mutual information (MI) between the learned representation and the input as a metric to find where in training, the network latches onto shortcuts. Experiments demonstrate that MI can be used as a domain-agnostic metric for monitoring shortcut learning.
Hospital-Agnostic Image Representation Learning in Digital Pathology
Apr 05, 2022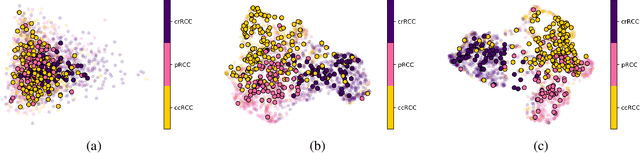
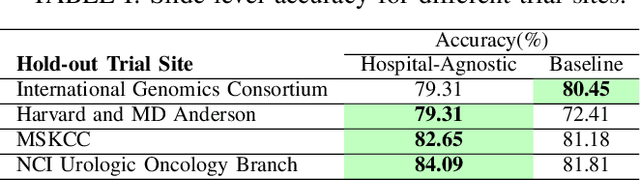
Abstract:Whole Slide Images (WSIs) in digital pathology are used to diagnose cancer subtypes. The difference in procedures to acquire WSIs at various trial sites gives rise to variability in the histopathology images, thus making consistent diagnosis challenging. These differences may stem from variability in image acquisition through multi-vendor scanners, variable acquisition parameters, and differences in staining procedure; as well, patient demographics may bias the glass slide batches before image acquisition. These variabilities are assumed to cause a domain shift in the images of different hospitals. It is crucial to overcome this domain shift because an ideal machine-learning model must be able to work on the diverse sources of images, independent of the acquisition center. A domain generalization technique is leveraged in this study to improve the generalization capability of a Deep Neural Network (DNN), to an unseen histopathology image set (i.e., from an unseen hospital/trial site) in the presence of domain shift. According to experimental results, the conventional supervised-learning regime generalizes poorly to data collected from different hospitals. However, the proposed hospital-agnostic learning can improve the generalization considering the low-dimensional latent space representation visualization, and classification accuracy results.
Learning to Predict RNA Sequence Expressions from Whole Slide Images with Applications for Search and Classification
Mar 26, 2022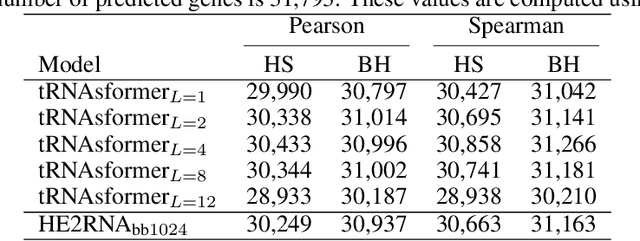
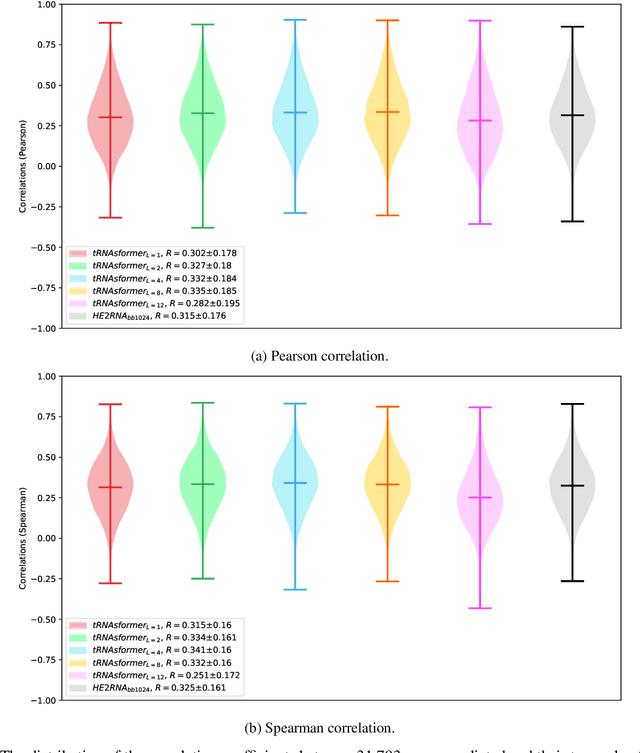
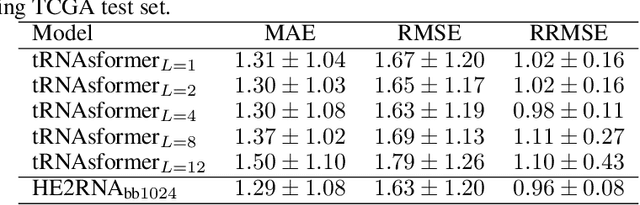
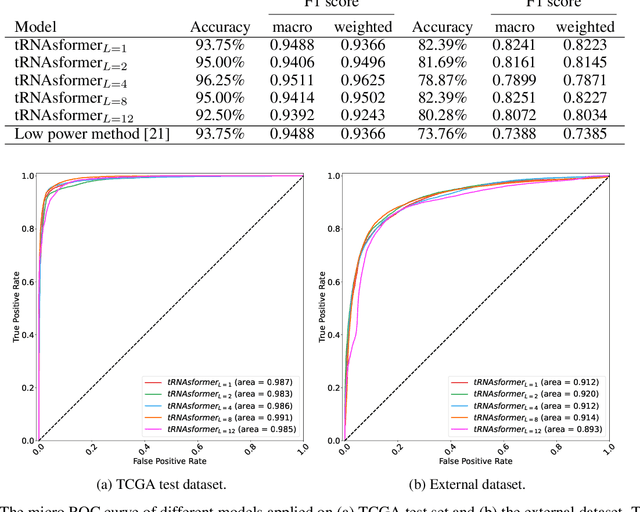
Abstract:Deep learning methods are widely applied in digital pathology to address clinical challenges such as prognosis and diagnosis. As one of the most recent applications, deep models have also been used to extract molecular features from whole slide images. Although molecular tests carry rich information, they are often expensive, time-consuming, and require additional tissue to sample. In this paper, we propose tRNAsfomer, an attention-based topology that can learn both to predict the bulk RNA-seq from an image and represent the whole slide image of a glass slide simultaneously. The tRNAsfomer uses multiple instance learning to solve a weakly supervised problem while the pixel-level annotation is not available for an image. We conducted several experiments and achieved better performance and faster convergence in comparison to the state-of-the-art algorithms. The proposed tRNAsfomer can assist as a computational pathology tool to facilitate a new generation of search and classification methods by combining the tissue morphology and the molecular fingerprint of the biopsy samples.
Gram Barcodes for Histopathology Tissue Texture Retrieval
Nov 28, 2021
Abstract:Recent advances in digital pathology have led to the need for Histopathology Image Retrieval (HIR) systems that search through databases of biopsy images to find similar cases to a given query image. These HIR systems allow pathologists to effortlessly and efficiently access thousands of previously diagnosed cases in order to exploit the knowledge in the corresponding pathology reports. Since HIR systems may have to deal with millions of gigapixel images, the extraction of compact and expressive image features must be available to allow for efficient and accurate retrieval. In this paper, we propose the application of Gram barcodes as image features for HIR systems. Unlike most feature generation schemes, Gram barcodes are based on high-order statistics that describe tissue texture by summarizing the correlations between different feature maps in layers of convolutional neural networks. We run HIR experiments on three public datasets using a pre-trained VGG19 network for Gram barcode generation and showcase highly competitive results.
Beyond Neighbourhood-Preserving Transformations for Quantization-Based Unsupervised Hashing
Oct 01, 2021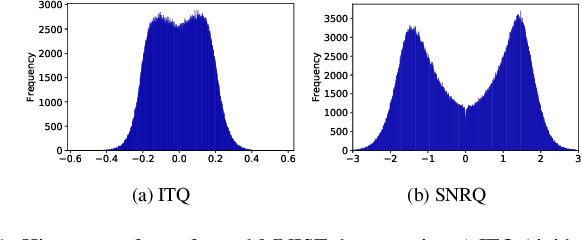
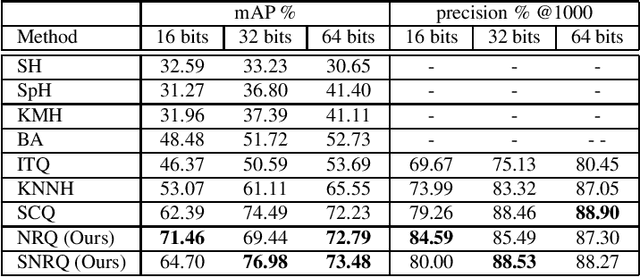
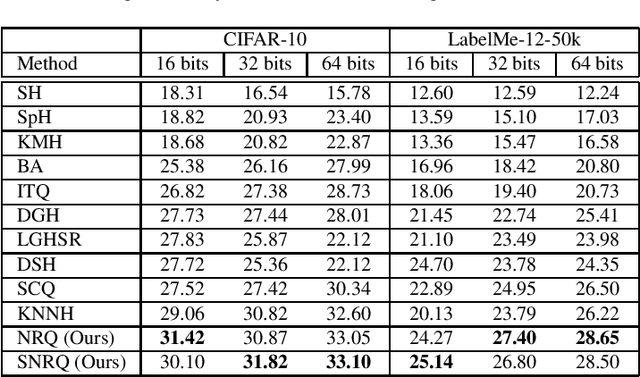
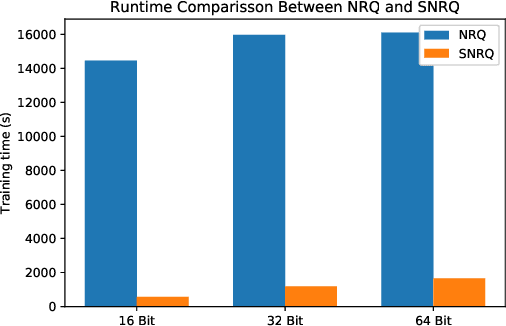
Abstract:An effective unsupervised hashing algorithm leads to compact binary codes preserving the neighborhood structure of data as much as possible. One of the most established schemes for unsupervised hashing is to reduce the dimensionality of data and then find a rigid (neighbourhood-preserving) transformation that reduces the quantization error. Although employing rigid transformations is effective, we may not reduce quantization loss to the ultimate limits. As well, reducing dimensionality and quantization loss in two separate steps seems to be sub-optimal. Motivated by these shortcomings, we propose to employ both rigid and non-rigid transformations to reduce quantization error and dimensionality simultaneously. We relax the orthogonality constraint on the projection in a PCA-formulation and regularize this by a quantization term. We show that both the non-rigid projection matrix and rotation matrix contribute towards minimizing quantization loss but in different ways. A scalable nested coordinate descent approach is proposed to optimize this mixed-integer optimization problem. We evaluate the proposed method on five public benchmark datasets providing almost half a million images. Comparative results indicate that the proposed method mostly outperforms state-of-art linear methods and competes with end-to-end deep solutions.
Unsupervised Detection of Lung Nodules in Chest Radiography Using Generative Adversarial Networks
Aug 04, 2021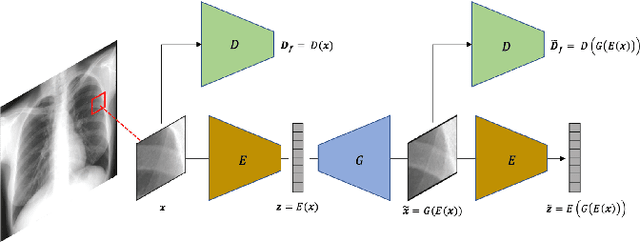
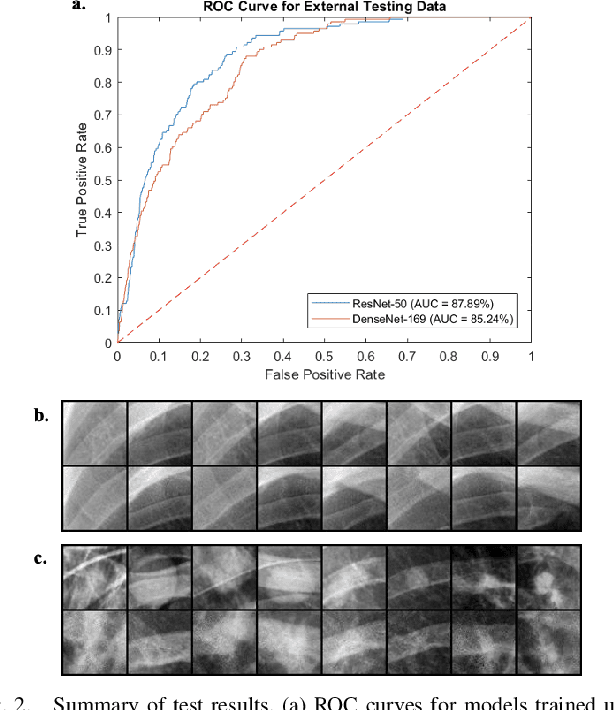
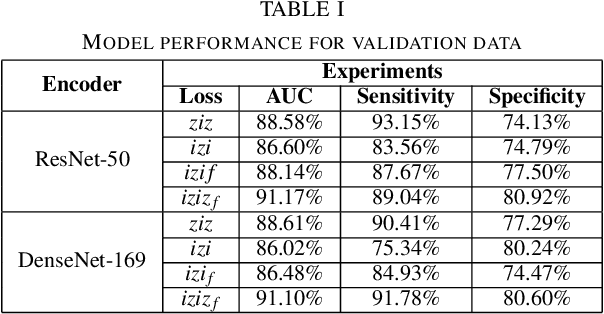
Abstract:Lung nodules are commonly missed in chest radiographs. We propose and evaluate P-AnoGAN, an unsupervised anomaly detection approach for lung nodules in radiographs. P-AnoGAN modifies the fast anomaly detection generative adversarial network (f-AnoGAN) by utilizing a progressive GAN and a convolutional encoder-decoder-encoder pipeline. Model training uses only unlabelled healthy lung patches extracted from the Indiana University Chest X-Ray Collection. External validation and testing are performed using healthy and unhealthy patches extracted from the ChestX-ray14 and Japanese Society for Radiological Technology datasets, respectively. Our model robustly identifies patches containing lung nodules in external validation and test data with ROC-AUC of 91.17% and 87.89%, respectively. These results show unsupervised methods may be useful in challenging tasks such as lung nodule detection in radiographs.
Automatic Multi-Stain Registration of Whole Slide Images in Histopathology
Jul 29, 2021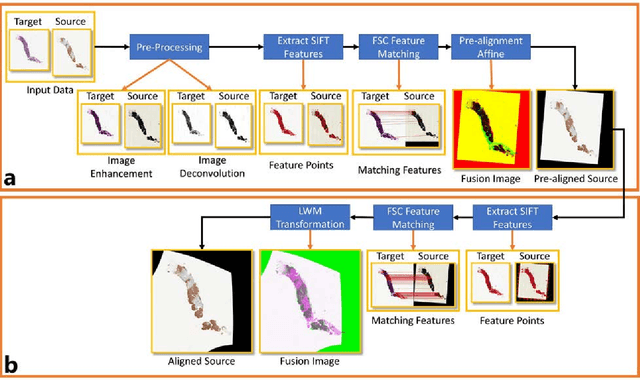
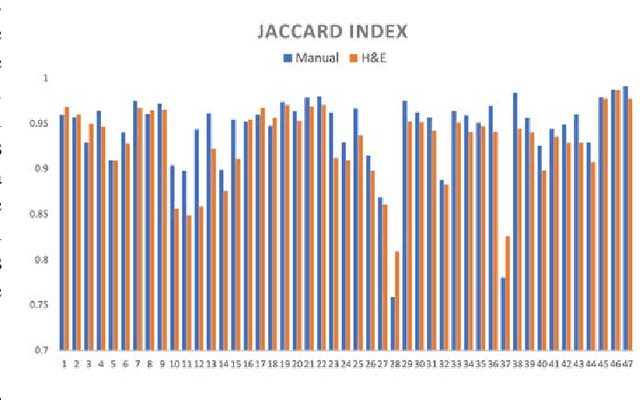
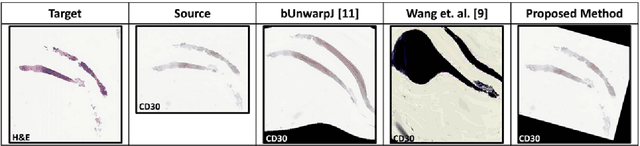
Abstract:Joint analysis of multiple biomarker images and tissue morphology is important for disease diagnosis, treatment planning and drug development. It requires cross-staining comparison among Whole Slide Images (WSIs) of immuno-histochemical and hematoxylin and eosin (H&E) microscopic slides. However, automatic, and fast cross-staining alignment of enormous gigapixel WSIs at single-cell precision is challenging. In addition to morphological deformations introduced during slide preparation, there are large variations in cell appearance and tissue morphology across different staining. In this paper, we propose a two-step automatic feature-based cross-staining WSI alignment to assist localization of even tiny metastatic foci in the assessment of lymph node. Image pairs were aligned allowing for translation, rotation, and scaling. The registration was performed automatically by first detecting landmarks in both images, using the scale-invariant image transform (SIFT), followed by the fast sample consensus (FSC) protocol for finding point correspondences and finally aligned the images. The Registration results were evaluated using both visual and quantitative criteria using the Jaccard index. The average Jaccard similarity index of the results produced by the proposed system is 0.942 when compared with the manual registration.
A Similarity Measure of Histopathology Images by Deep Embeddings
Jul 29, 2021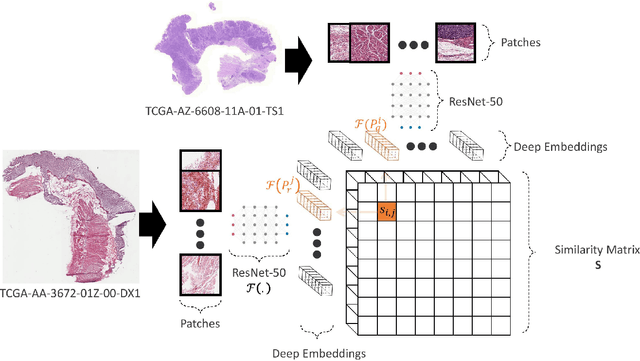
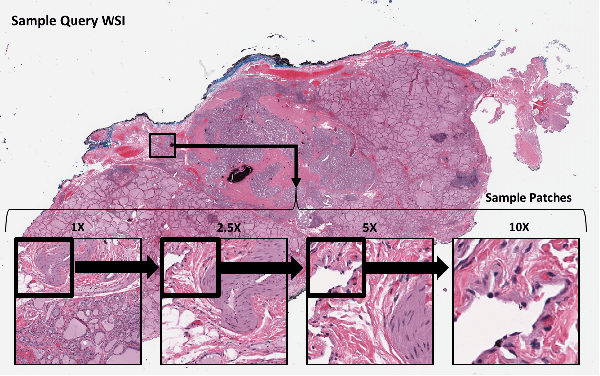

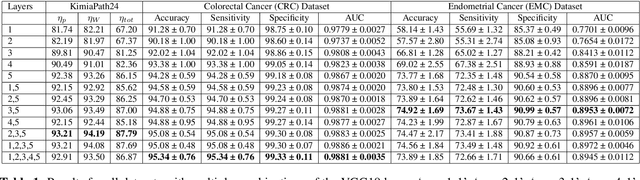
Abstract:Histopathology digital scans are large-size images that contain valuable information at the pixel level. Content-based comparison of these images is a challenging task. This study proposes a content-based similarity measure for high-resolution gigapixel histopathology images. The proposed similarity measure is an expansion of cosine vector similarity to a matrix. Each image is divided into same-size patches with a meaningful amount of information (i.e., contained enough tissue). The similarity is measured by the extraction of patch-level deep embeddings of the last pooling layer of a pre-trained deep model at four different magnification levels, namely, 1x, 2.5x, 5x, and 10x magnifications. In addition, for faster measurement, embedding reduction is investigated. Finally, to assess the proposed method, an image search method is implemented. Results show that the similarity measure represents the slide labels with a maximum accuracy of 93.18\% for top-5 search at 5x magnification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge