Gladston Moreira
Benford's Law as a Distributional Prior for Post-Training Quantization of Large Language Models
Jan 29, 2026Abstract:The rapid growth of Large Language Models (LLMs) intensifies the need for effective compression, with weight quantization being the most widely adopted technique. Standard uniform quantizers assume that parameters are evenly distributed, an assumption at odds with the highly skewed distributions observed in practice. We propose Benford-Quant, a simple, data-free non-uniform quantizer inspired by Benford's Law, which predicts that leading digits follow a logarithmic distribution. Benford-Quant replaces the uniform grid with a log-spaced codebook, dedicating more resolution to the frequent small-magnitude weights. We provide both theoretical intuition and empirical evidence: (i) weights in transformer transformational layers adhere closely to Benford statistics, while normalization layers systematically deviate; (ii) on Small Language Models (SLMs), Benford-Quant consistently improves perplexity, reducing 4-bit perplexity on Gemma-270M by more than 10%; and (iii) on larger LLMs, it remains competitive, with differences explained by over-parameterization effects. Our results indicate that incorporating a Benford-inspired prior into quantization grids is a low-cost modification that yields accuracy gains in aggressive few-bit regimes. Although it is not able to surpass the state of the art in tasks such as perplexity and LAMBADA, the Benford-Quant approach can be hybridized with other quantization methods-such as SmoothQuant and Activation-Aware Quantization-without major pipeline modification, potentially improving their performance.
PD-Loss: Proxy-Decidability for Efficient Metric Learning
Aug 23, 2025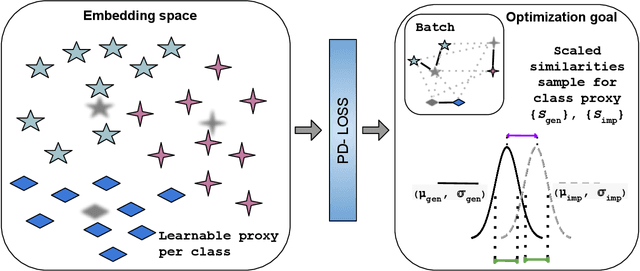
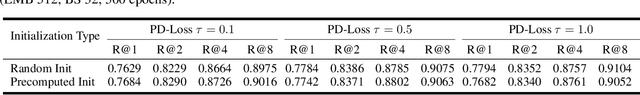
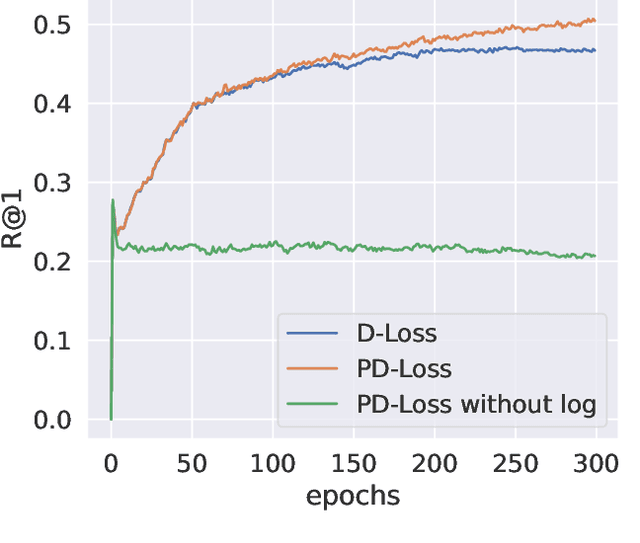
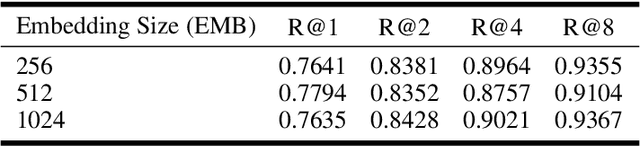
Abstract:Deep Metric Learning (DML) aims to learn embedding functions that map semantically similar inputs to proximate points in a metric space while separating dissimilar ones. Existing methods, such as pairwise losses, are hindered by complex sampling requirements and slow convergence. In contrast, proxy-based losses, despite their improved scalability, often fail to optimize global distribution properties. The Decidability-based Loss (D-Loss) addresses this by targeting the decidability index (d') to enhance distribution separability, but its reliance on large mini-batches imposes significant computational constraints. We introduce Proxy-Decidability Loss (PD-Loss), a novel objective that integrates learnable proxies with the statistical framework of d' to optimize embedding spaces efficiently. By estimating genuine and impostor distributions through proxies, PD-Loss combines the computational efficiency of proxy-based methods with the principled separability of D-Loss, offering a scalable approach to distribution-aware DML. Experiments across various tasks, including fine-grained classification and face verification, demonstrate that PD-Loss achieves performance comparable to that of state-of-the-art methods while introducing a new perspective on embedding optimization, with potential for broader applications.
Enhancing Decision Space Diversity in Multi-Objective Evolutionary Optimization for the Diet Problem
Aug 09, 2025Abstract:Multi-objective evolutionary algorithms (MOEAs) are essential for solving complex optimization problems, such as the diet problem, where balancing conflicting objectives, like cost and nutritional content, is crucial. However, most MOEAs focus on optimizing solutions in the objective space, often neglecting the diversity of solutions in the decision space, which is critical for providing decision-makers with a wide range of choices. This paper introduces an approach that directly integrates a Hamming distance-based measure of uniformity into the selection mechanism of a MOEA to enhance decision space diversity. Experiments on a multi-objective formulation of the diet problem demonstrate that our approach significantly improves decision space diversity compared to NSGA-II, while maintaining comparable objective space performance. The proposed method offers a generalizable strategy for integrating decision space awareness into MOEAs.
Investigating the Impact of Large-Scale Pre-training on Nutritional Content Estimation from 2D Images
Aug 06, 2025



Abstract:Estimating the nutritional content of food from images is a critical task with significant implications for health and dietary monitoring. This is challenging, especially when relying solely on 2D images, due to the variability in food presentation, lighting, and the inherent difficulty in inferring volume and mass without depth information. Furthermore, reproducibility in this domain is hampered by the reliance of state-of-the-art methods on proprietary datasets for large-scale pre-training. In this paper, we investigate the impact of large-scale pre-training datasets on the performance of deep learning models for nutritional estimation using only 2D images. We fine-tune and evaluate Vision Transformer (ViT) models pre-trained on two large public datasets, ImageNet and COYO, comparing their performance against baseline CNN models (InceptionV2 and ResNet-50) and a state-of-the-art method pre-trained on the proprietary JFT-300M dataset. We conduct extensive experiments on the Nutrition5k dataset, a large-scale collection of real-world food plates with high-precision nutritional annotations. Our evaluation using Mean Absolute Error (MAE) and Mean Absolute Percentage Error (MAE%) reveals that models pre-trained on JFT-300M significantly outperform those pre-trained on public datasets. Unexpectedly, the model pre-trained on the massive COYO dataset performs worse than the model pre-trained on ImageNet for this specific regression task, refuting our initial hypothesis. Our analysis provides quantitative evidence highlighting the critical role of pre-training dataset characteristics, including scale, domain relevance, and curation quality, for effective transfer learning in 2D nutritional estimation.
A Systematic Review of ECG Arrhythmia Classification: Adherence to Standards, Fair Evaluation, and Embedded Feasibility
Mar 10, 2025



Abstract:The classification of electrocardiogram (ECG) signals is crucial for early detection of arrhythmias and other cardiac conditions. However, despite advances in machine learning, many studies fail to follow standardization protocols, leading to inconsistencies in performance evaluation and real-world applicability. Additionally, hardware constraints essential for practical deployment, such as in pacemakers, Holter monitors, and wearable ECG patches, are often overlooked. Since real-world impact depends on feasibility in resource-constrained devices, ensuring efficient deployment is critical for continuous monitoring. This review systematically analyzes ECG classification studies published between 2017 and 2024, focusing on those adhering to the E3C (Embedded, Clinical, and Comparative Criteria), which include inter-patient paradigm implementation, compliance with Association for the Advancement of Medical Instrumentation (AAMI) recommendations, and model feasibility for embedded systems. While many studies report high accuracy, few properly consider patient-independent partitioning and hardware limitations. We identify state-of-the-art methods meeting E3C criteria and conduct a comparative analysis of accuracy, inference time, energy consumption, and memory usage. Finally, we propose standardized reporting practices to ensure fair comparisons and practical applicability of ECG classification models. By addressing these gaps, this study aims to guide future research toward more robust and clinically viable ECG classification systems.
CapsProm: A Capsule Network For Promoter Prediction
Dec 07, 2021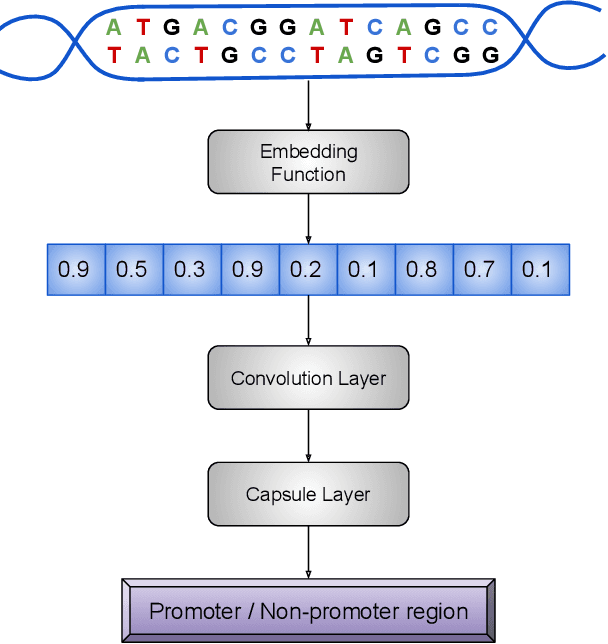
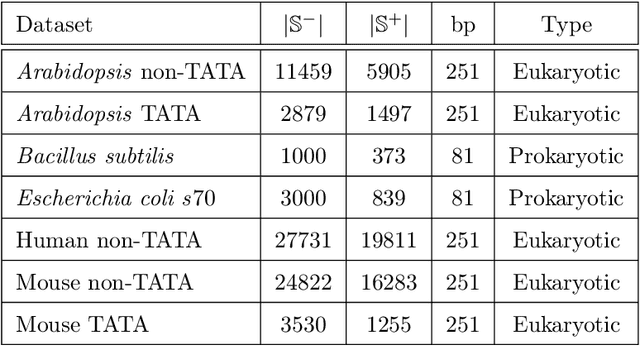
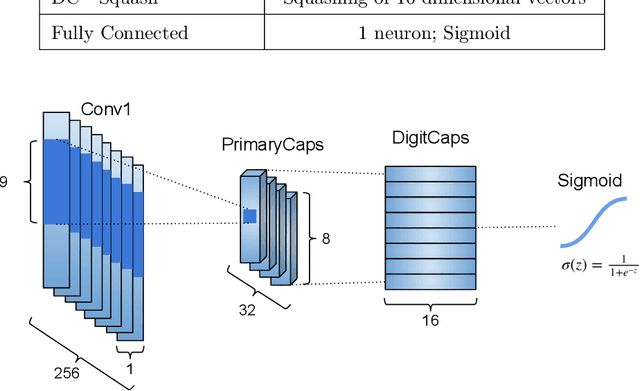
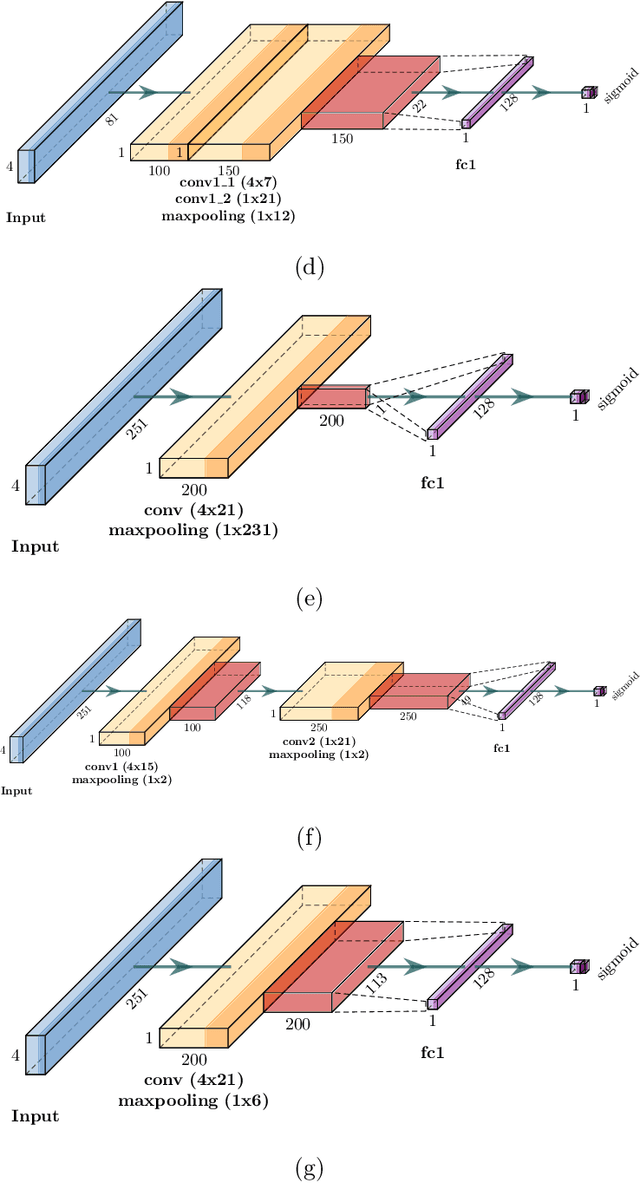
Abstract:Locating the promoter region in DNA sequences is of paramount importance in the field of bioinformatics. This is a problem widely studied in the literature, however, not yet fully resolved. Some researchers have presented remarkable results using convolution networks, that allowed the automatic extraction of features from a DNA chain. However, a universal architecture that could generalize to several organisms has not yet been achieved, and thus, requiring researchers to seek new architectures and hyperparameters for each new organism evaluated. In this work, we propose a versatile architecture, based on capsule network, that can accurately identify promoter sequences in raw DNA data from seven different organisms, eukaryotic, and prokaryotic. Our model, the CapsProm, could assist in the transfer of learning between organisms and expand its applicability. Furthermore the CapsProm showed competitive results, overcoming the baseline method in five out of seven of the tested datasets (F1-score). The models and source code are made available at https://github.com/lauromoraes/CapsNet-promoter.
A Decidability-Based Loss Function
Sep 12, 2021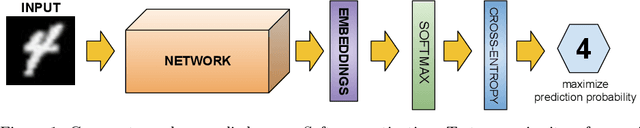
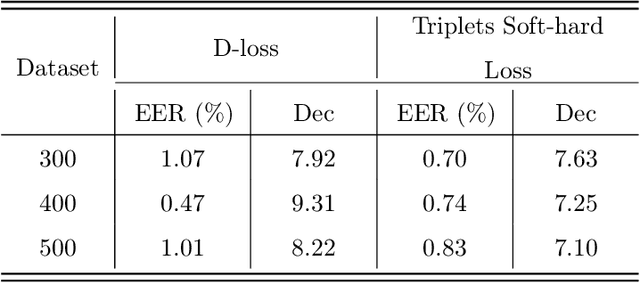
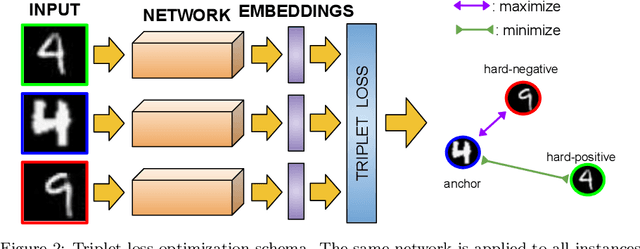
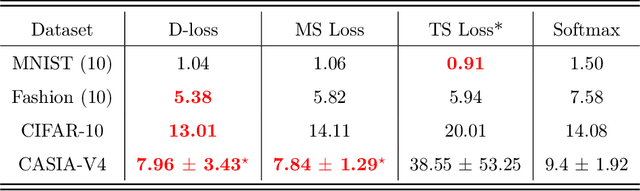
Abstract:Nowadays, deep learning is the standard approach for a wide range of problems, including biometrics, such as face recognition and speech recognition, etc. Biometric problems often use deep learning models to extract features from images, also known as embeddings. Moreover, the loss function used during training strongly influences the quality of the generated embeddings. In this work, a loss function based on the decidability index is proposed to improve the quality of embeddings for the verification routine. Our proposal, the D-loss, avoids some Triplet-based loss disadvantages such as the use of hard samples and tricky parameter tuning, which can lead to slow convergence. The proposed approach is compared against the Softmax (cross-entropy), Triplets Soft-Hard, and the Multi Similarity losses in four different benchmarks: MNIST, Fashion-MNIST, CIFAR10 and CASIA-IrisV4. The achieved results show the efficacy of the proposal when compared to other popular metrics in the literature. The D-loss computation, besides being simple, non-parametric and easy to implement, favors both the inter-class and intra-class scenarios.
Towards an Effective and Efficient Deep Learning Model for COVID-19 Patterns Detection in X-ray Images
Apr 28, 2020
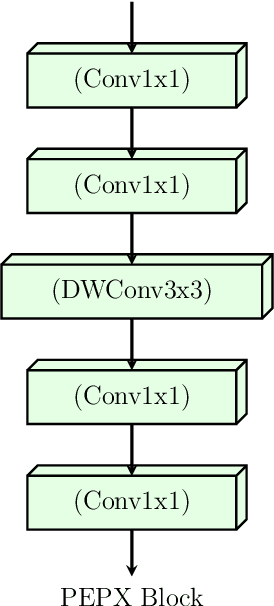
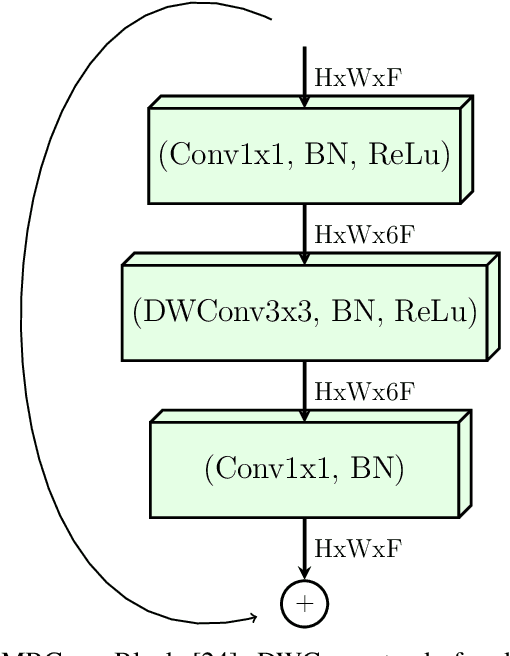
Abstract:Confronting the pandemic of COVID-19 caused by the new coronavirus, the SARS-CoV-2, is nowadays one of the most prominent challenges of the human species. A key factor in slowing down the virus propagation is the rapid diagnosis and isolation of infected patients. Nevertheless, the standard method for COVID-19 identification, the Reverse transcription polymerase chain reaction (RT-PCR) method, is time-consuming and in short supply due to the pandemic. Researchers around the world have been looking for alternative screening methods. In this context, deep learning applied to chest X-rays of patients has been showing promising results in the identification of COVID-19. Despite their success, the computational cost of these methods remains high, which imposes difficulties in their accessibility and availability. Thus, in this work, we propose to explore and extend the EfficientNet family of models using chest X-rays images to perform COVID-19 detection. As a result, we can produce a high-quality model with an overall accuracy of 93.9%, COVID-19, sensitivity of 96.8% and positive prediction of 100% while having about 30 times fewer parameters than the baseline literature model, 28 and 5 times fewer parameters than the popular VGG16 and ResNet50 architectures, respectively. We believe the reported figures represent state-of-the-art results, both in terms of efficiency and effectiveness, for the COVIDx database, a database comprised of 13,800 X-ray images, 183 of which are from patients affected by COVID-19.
Simultaneous Iris and Periocular Region Detection Using Coarse Annotations
Jul 31, 2019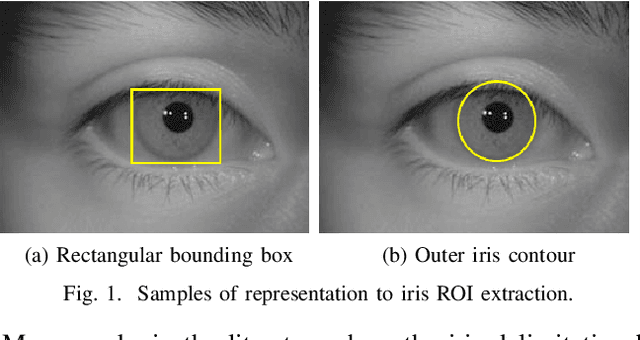
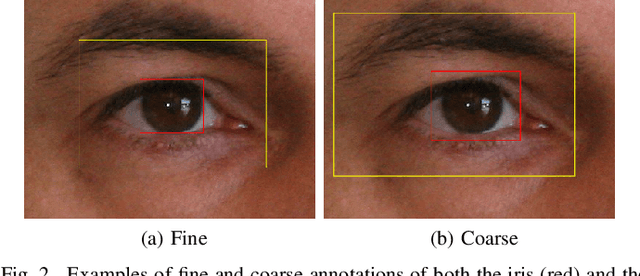
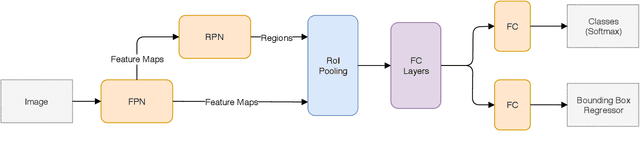
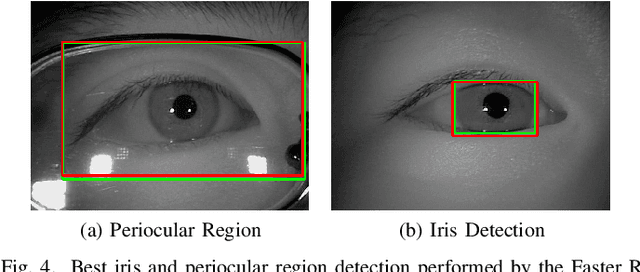
Abstract:In this work, we propose to detect the iris and periocular regions simultaneously using coarse annotations and two well-known object detectors: YOLOv2 and Faster R-CNN. We believe coarse annotations can be used in recognition systems based on the iris and periocular regions, given the much smaller engineering effort required to manually annotate the training images. We manually made coarse annotations of the iris and periocular regions (122K images from the visible (VIS) spectrum and 38K images from the near-infrared (NIR) spectrum). The iris annotations in the NIR databases were generated semi-automatically by first applying an iris segmentation CNN and then performing a manual inspection. These annotations were made for 11 well-known public databases (3 NIR and 8 VIS) designed for the iris-based recognition problem and are publicly available to the research community. Experimenting our proposal on these databases, we highlight two results. First, the Faster R-CNN + Feature Pyramid Network (FPN) model reported an Intersection over Union (IoU) higher than YOLOv2 (91.86% vs 85.30%). Second, the detection of the iris and periocular regions being performed simultaneously is as accurate as performed separately, but with a lower computational cost, i.e., two tasks were carried out at the cost of one.
A Benchmark for Iris Location and a Deep Learning Detector Evaluation
Apr 30, 2018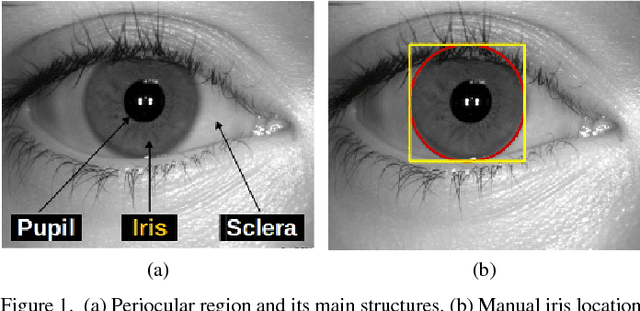

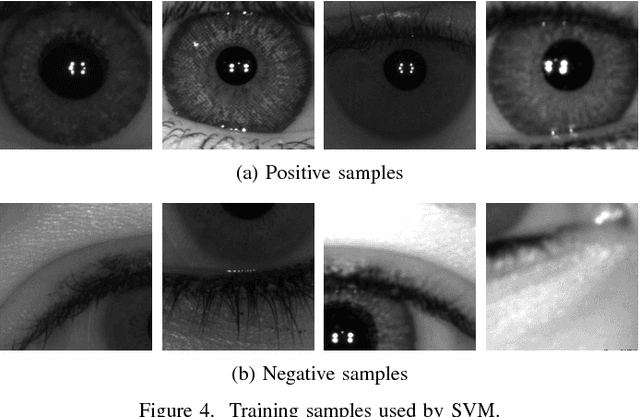
Abstract:The iris is considered as the biometric trait with the highest unique probability. The iris location is an important task for biometrics systems, affecting directly the results obtained in specific applications such as iris recognition, spoofing and contact lenses detection, among others. This work defines the iris location problem as the delimitation of the smallest squared window that encompasses the iris region. In order to build a benchmark for iris location we annotate (iris squared bounding boxes) four databases from different biometric applications and make them publicly available to the community. Besides these 4 annotated databases, we include 2 others from the literature. We perform experiments on these six databases, five obtained with near infra-red sensors and one with visible light sensor. We compare the classical and outstanding Daugman iris location approach with two window based detectors: 1) a sliding window detector based on features from Histogram of Oriented Gradients (HOG) and a linear Support Vector Machines (SVM) classifier; 2) a deep learning based detector fine-tuned from YOLO object detector. Experimental results showed that the deep learning based detector outperforms the other ones in terms of accuracy and runtime (GPUs version) and should be chosen whenever possible.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge