Frederik L. Dennig
LCIP: Loss-Controlled Inverse Projection of High-Dimensional Image Data
Feb 11, 2026Abstract:Projections (or dimensionality reduction) methods $P$ aim to map high-dimensional data to typically 2D scatterplots for visual exploration. Inverse projection methods $P^{-1}$ aim to map this 2D space to the data space to support tasks such as data augmentation, classifier analysis, and data imputation. Current $P^{-1}$ methods suffer from a fundamental limitation -- they can only generate a fixed surface-like structure in data space, which poorly covers the richness of this space. We address this by a new method that can `sweep' the data space under user control. Our method works generically for any $P$ technique and dataset, is controlled by two intuitive user-set parameters, and is simple to implement. We demonstrate it by an extensive application involving image manipulation for style transfer.
Evaluating Autoencoders for Parametric and Invertible Multidimensional Projections
Apr 23, 2025



Abstract:Recently, neural networks have gained attention for creating parametric and invertible multidimensional data projections. Parametric projections allow for embedding previously unseen data without recomputing the projection as a whole, while invertible projections enable the generation of new data points. However, these properties have never been explored simultaneously for arbitrary projection methods. We evaluate three autoencoder (AE) architectures for creating parametric and invertible projections. Based on a given projection, we train AEs to learn a mapping into 2D space and an inverse mapping into the original space. We perform a quantitative and qualitative comparison on four datasets of varying dimensionality and pattern complexity using t-SNE. Our results indicate that AEs with a customized loss function can create smoother parametric and inverse projections than feed-forward neural networks while giving users control over the strength of the smoothing effect.
FDive: Learning Relevance Models using Pattern-based Similarity Measures
Jul 30, 2019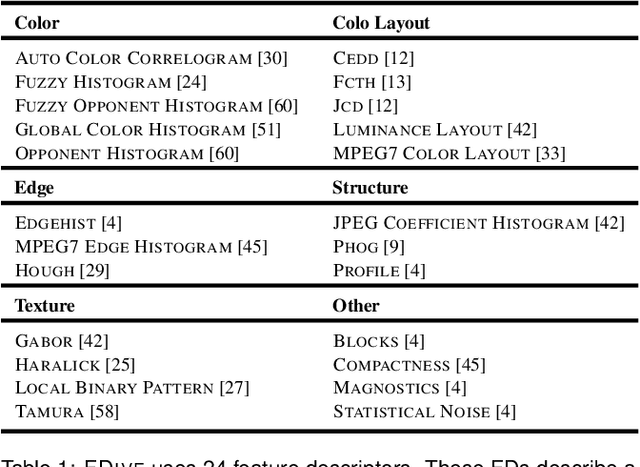
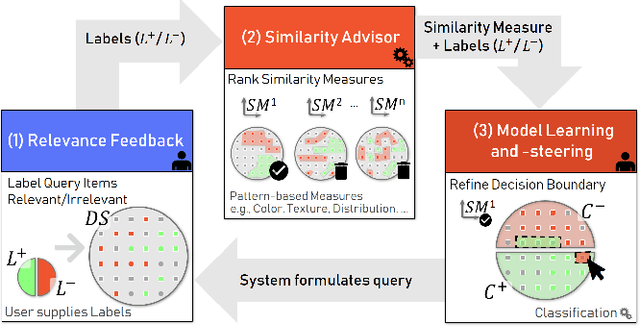
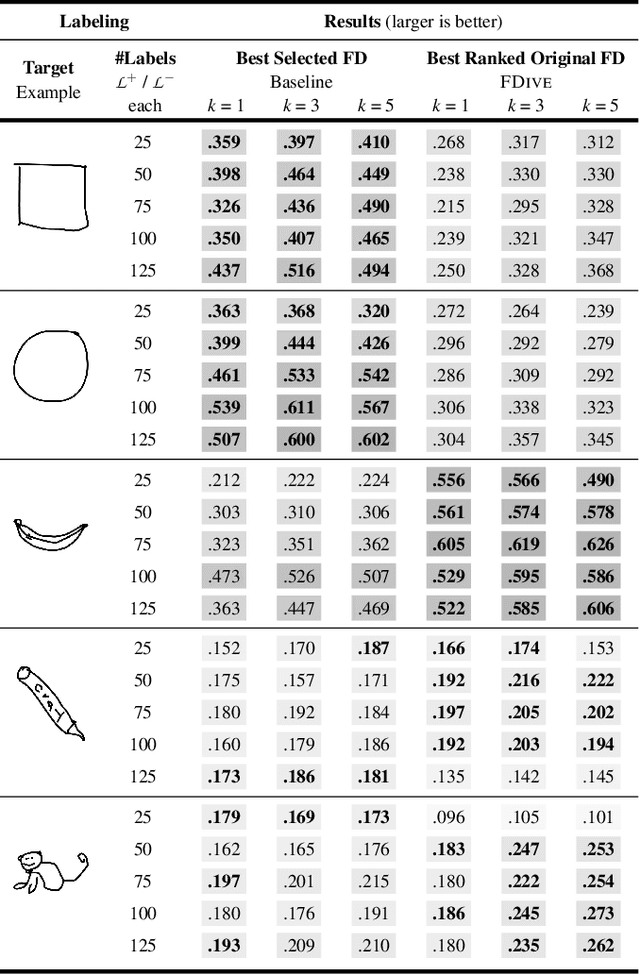
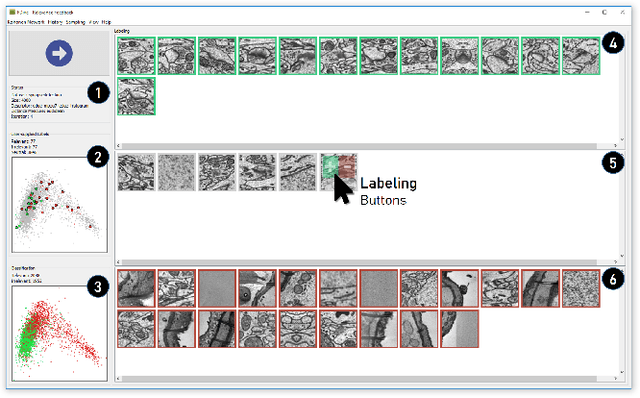
Abstract:The detection of interesting patterns in large high-dimensional datasets is difficult because of their dimensionality and pattern complexity. Therefore, analysts require automated support for the extraction of relevant patterns. In this paper, we present FDive, a visual active learning system that helps to create visually explorable relevance models, assisted by learning a pattern-based similarity. We use a small set of user-provided labels to rank similarity measures, consisting of feature descriptor and distance function combinations, by their ability to distinguish relevant from irrelevant data. Based on the best-ranked similarity measure, the system calculates an interactive Self-Organizing Map-based relevance model, which classifies data according to the cluster affiliation. It also automatically prompts further relevance feedback to improve its accuracy. Uncertain areas, especially near the decision boundaries, are highlighted and can be refined by the user. We evaluate our approach by comparison to state-of-the-art feature selection techniques and demonstrate the usefulness of our approach by a case study classifying electron microscopy images of brain cells. The results show that FDive enhances both the quality and understanding of relevance models and can thus lead to new insights for brain research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge