Dmitry V. Dylov
Tubular Shape Aware Data Generation for Semantic Segmentation in Medical Imaging
Oct 02, 2020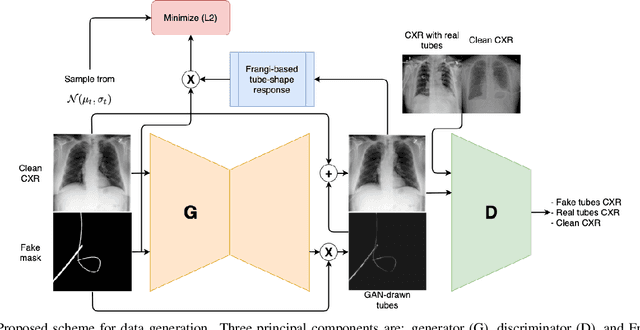
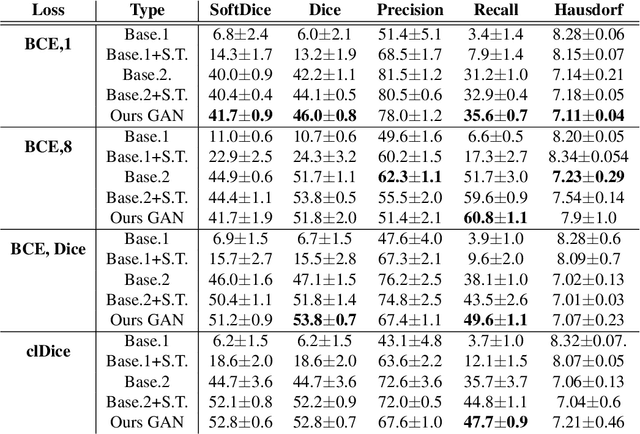
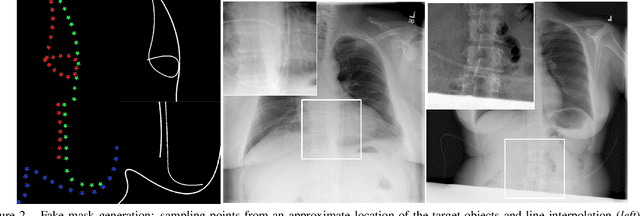
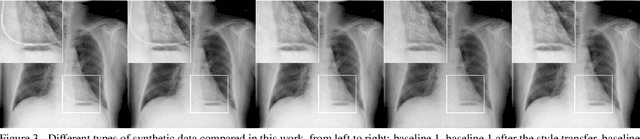
Abstract:Chest X-ray is one of the most widespread examinations of the human body. In interventional radiology, its use is frequently associated with the need to visualize various tube-like objects, such as puncture needles, guiding sheaths, wires, and catheters. Detection and precise localization of these tube-like objects in the X-ray images is, therefore, of utmost value, catalyzing the development of accurate target-specific segmentation algorithms. Similar to the other medical imaging tasks, the manual pixel-wise annotation of the tubes is a resource-consuming process. In this work, we aim to alleviate the lack of the annotated images by using artificial data. Specifically, we present an approach for synthetic data generation of the tube-shaped objects, with a generative adversarial network being regularized with a prior-shape constraint. Our method eliminates the need for paired image--mask data and requires only a weakly-labeled dataset (10--20 images) to reach the accuracy of the fully-supervised models. We report the applicability of the approach for the task of segmenting tubes and catheters in the X-ray images, whereas the results should also hold for the other imaging modalities.
Learnable Hollow Kernels for Anatomical Segmentation
Jul 09, 2020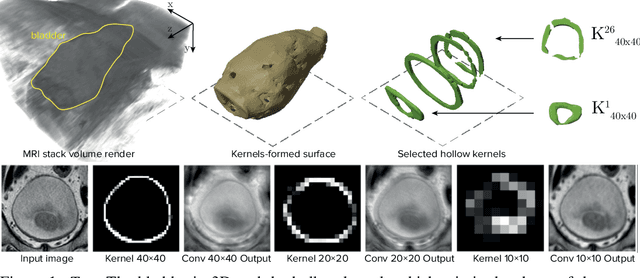
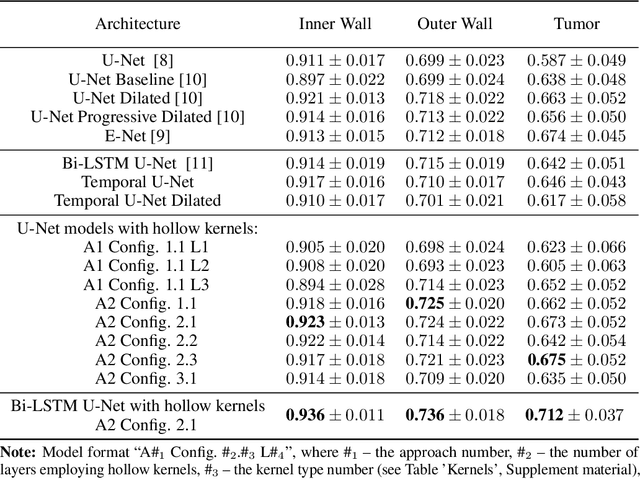
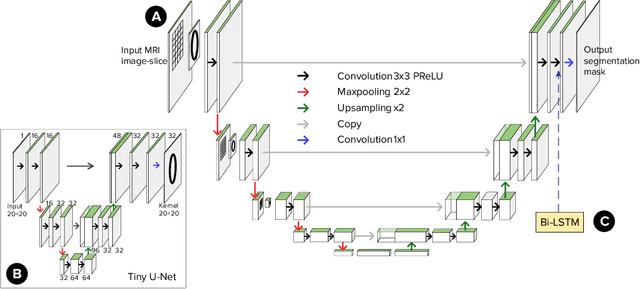
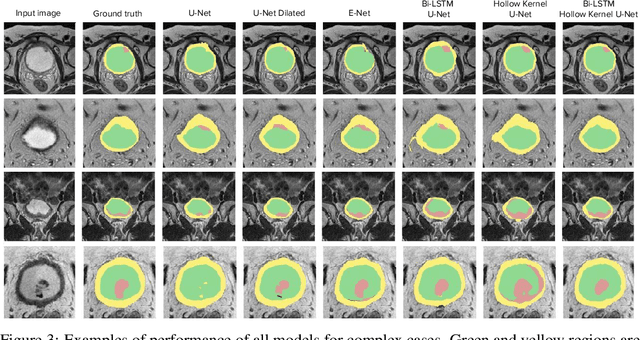
Abstract:Segmentation of certain hollow organs, such as the bladder, is especially hard to automate due to their complex geometry, vague intensity gradients in the soft tissues, and a tedious manual process of the data annotation routine. Yet, accurate localization of the walls and the cancer regions in the radiologic images of such organs is an essential step in oncology. To address this issue, we propose a new class of hollow kernels that learn to 'mimic' the contours of the segmented organ, effectively replicating its shape and structural complexity. We train a series of the U-Net-like neural networks using the proposed kernels and demonstrate the superiority of the idea in various spatio-temporal convolution scenarios. Specifically, the dilated hollow-kernel architecture outperforms state-of-the-art spatial segmentation models, whereas the addition of temporal blocks with, e.g., Bi-LSTM, establishes a new multi-class baseline for the bladder segmentation challenge. Our spatio-temporal model based on the hollow kernels reaches the mean dice scores of 0.936, 0.736, and 0.712 for the bladder's inner wall, the outer wall, and the tumor regions, respectively. The results pave the way towards other domain-specific deep learning applications where the shape of the segmented object could be used to form a proper convolution kernel for boosting the segmentation outcome.
Anomaly Detection with Deep Perceptual Autoencoders
Jun 23, 2020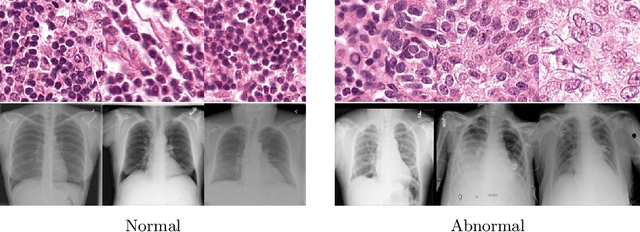
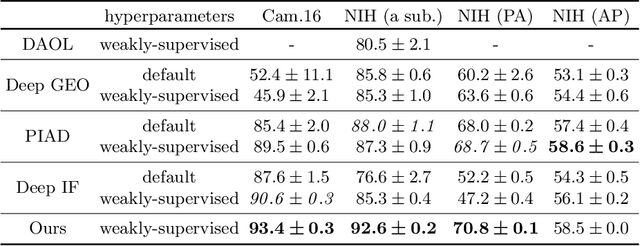
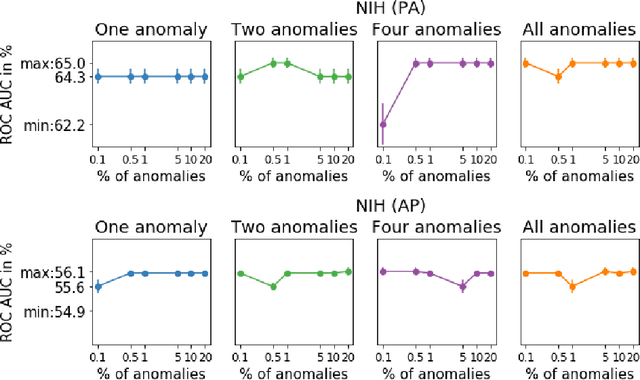

Abstract:Anomaly detection is the problem of recognizing abnormal inputs based on the seen examples of normal data. Despite recent advances of deep learning in recognizing image anomalies, these methods still prove incapable of handling complex medical images, such as barely visible abnormalities in chest X-rays and metastases in lymph nodes. To address this problem, we introduce a new powerful method of image anomaly detection. It relies on the classical autoencoder approach with a re-designed training pipeline to handle high-resolution, complex images and a robust way of computing an image abnormality score. We revisit the very problem statement of fully unsupervised anomaly detection, where no abnormal examples at all are provided during the model setup. We propose to relax this unrealistic assumption by using a very small number of anomalies of confined variability merely to initiate the search of hyperparameters of the model. We evaluate our solution on natural image datasets with a known benchmark, as well as on two medical datasets containing radiology and digital pathology images. The proposed approach suggests a new strong baseline for image anomaly detection and outperforms state-of-the-art approaches in complex medical image analysis tasks.
Deep Negative Volume Segmentation
Jun 22, 2020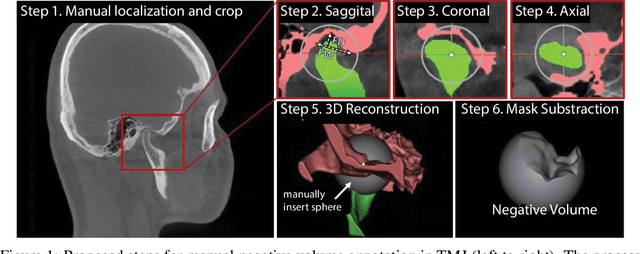
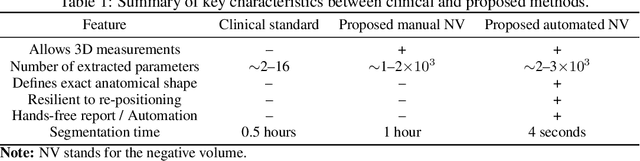
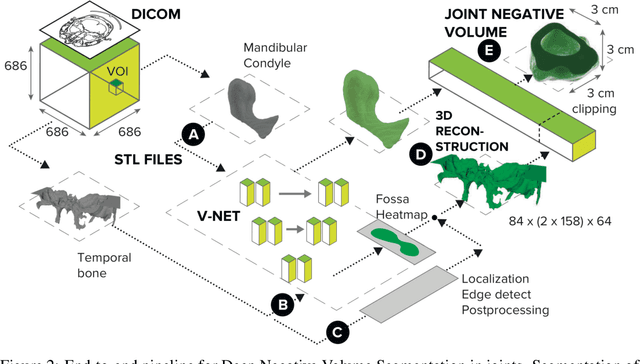
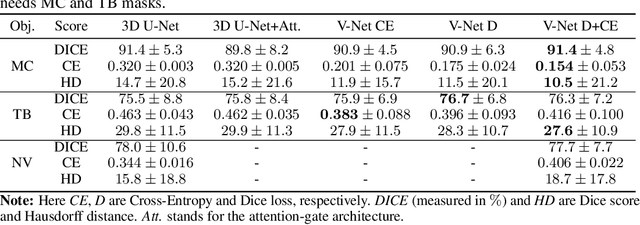
Abstract:Clinical examination of three-dimensional image data of compound anatomical objects, such as complex joints, remains a tedious process, demanding the time and the expertise of physicians. For instance, automation of the segmentation task of the TMJ (temporomandibular joint) has been hindered by its compound three-dimensional shape, multiple overlaid textures, an abundance of surrounding irregularities in the skull, and a virtually omnidirectional range of the jaw's motion - all of which extend the manual annotation process to more than an hour per patient. To address the challenge, we invent a new angle to the 3D segmentation task: namely, we propose to segment empty spaces between all the tissues surrounding the object - the so-called negative volume segmentation. Our approach is an end-to-end pipeline that comprises a V-Net for bone segmentation, a 3D volume construction by inflation of the reconstructed bone head in all directions along the normal vector to its mesh faces. Eventually confined within the skull bones, the inflated surface occupies the entire "negative" space in the joint, effectively providing a geometrical/topological metric of the joint's health. We validate the idea on the CT scans in a 50-patient dataset, annotated by experts in maxillofacial medicine, quantitatively compare the asymmetry given the left and the right negative volumes, and automate the entire framework for clinical adoption.
BRULÉ: Barycenter-Regularized Unsupervised Landmark Extraction
Jun 20, 2020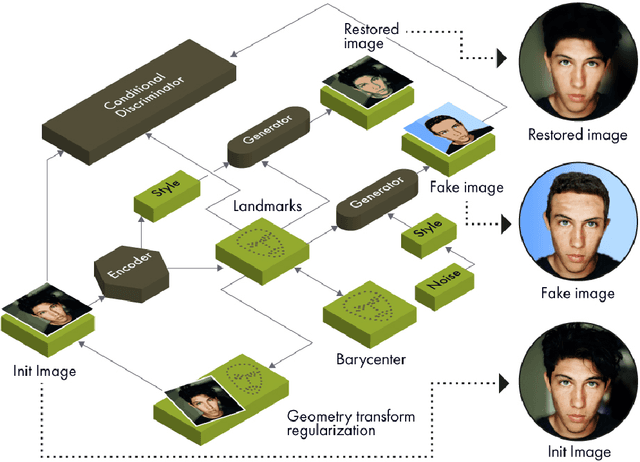
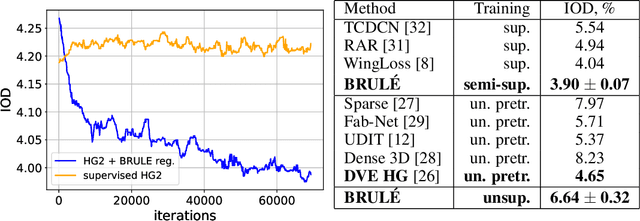
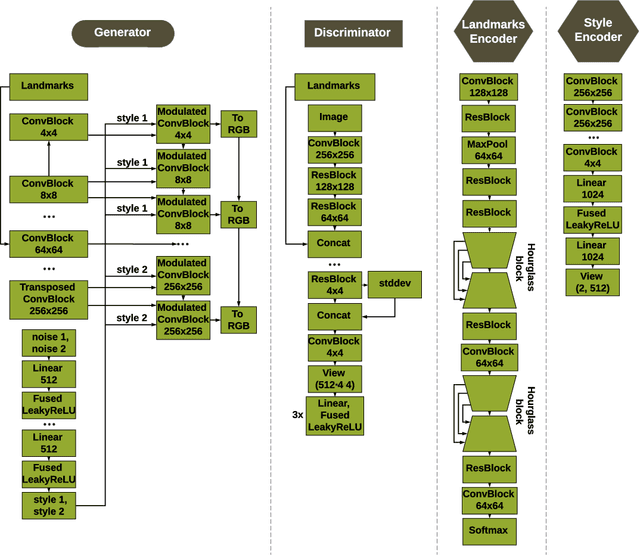

Abstract:Unsupervised retrieval of image features is vital for many computer vision tasks where the annotation is missing or scarce. In this work, we propose a new unsupervised approach to detect the landmarks in images, and we validate it on the popular task of human face key-points extraction. The method is based on the idea of auto-encoding the wanted landmarks in the latent space while discarding the non-essential information in the image and effectively preserving the interpretability. The interpretable latent space representation is achieved with the aid of a novel two-step regularization paradigm. The first regularization step evaluates transport distance from a given set of landmarks to the average value (the barycenter by Wasserstein distance). The second regularization step controls deviations from the barycenter by applying random geometric deformations synchronously to the initial image and to the encoded landmarks. During decoding, we add style features generated from the noise and reconstruct the initial image by the generative adversarial network (GAN) with transposed convolutions modulated by this style. We demonstrate the effectiveness of the approach both in unsupervised and in semi-supervised training scenarios using the 300-W and the CelebA datasets. The proposed regularization paradigm is shown to prevent overfitting, and the detection quality is shown to improve beyond the supervised outcome.
Reinforcement Learning Framework for Deep Brain Stimulation Study
Feb 22, 2020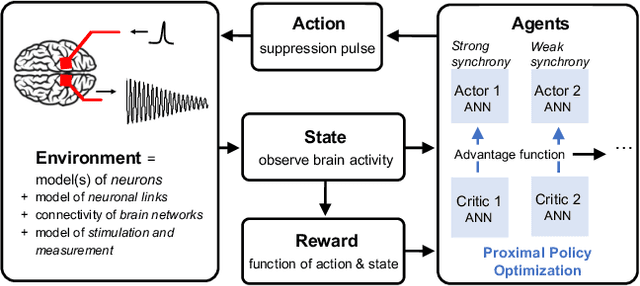
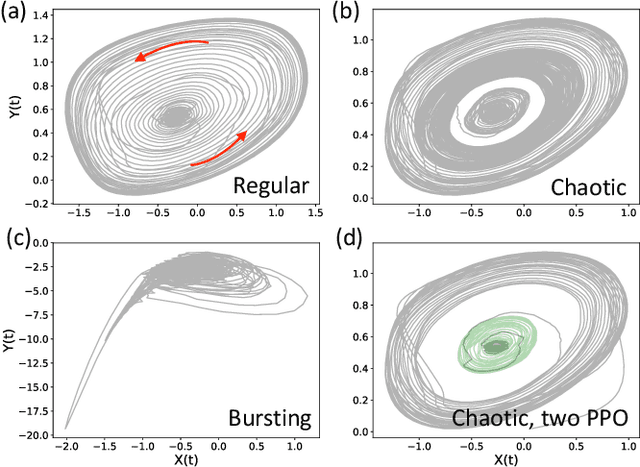
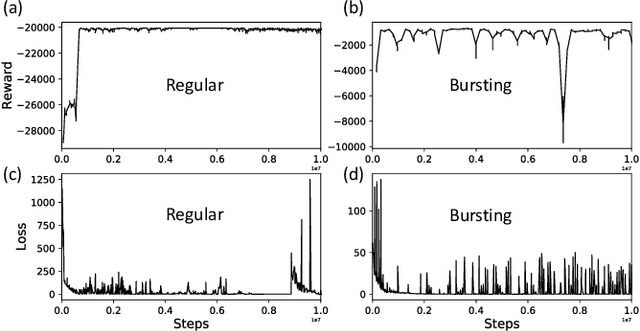
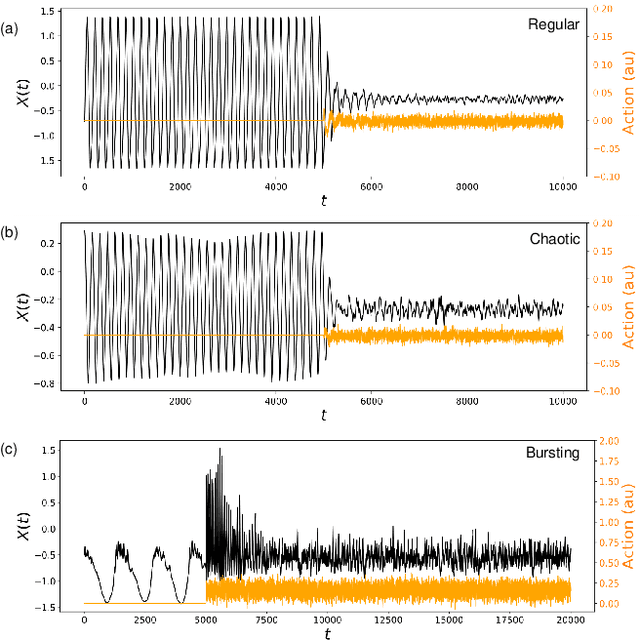
Abstract:Malfunctioning neurons in the brain sometimes operate synchronously, reportedly causing many neurological diseases, e.g. Parkinson's. Suppression and control of this collective synchronous activity are therefore of great importance for neuroscience, and can only rely on limited engineering trials due to the need to experiment with live human brains. We present the first Reinforcement Learning gym framework that emulates this collective behavior of neurons and allows us to find suppression parameters for the environment of synthetic degenerate models of neurons. We successfully suppress synchrony via RL for three pathological signaling regimes, characterize the framework's stability to noise, and further remove the unwanted oscillations by engaging multiple PPO agents.
Unsupervised non-parametric change point detection in quasi-periodic signals
Feb 07, 2020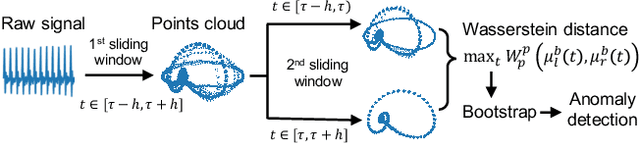
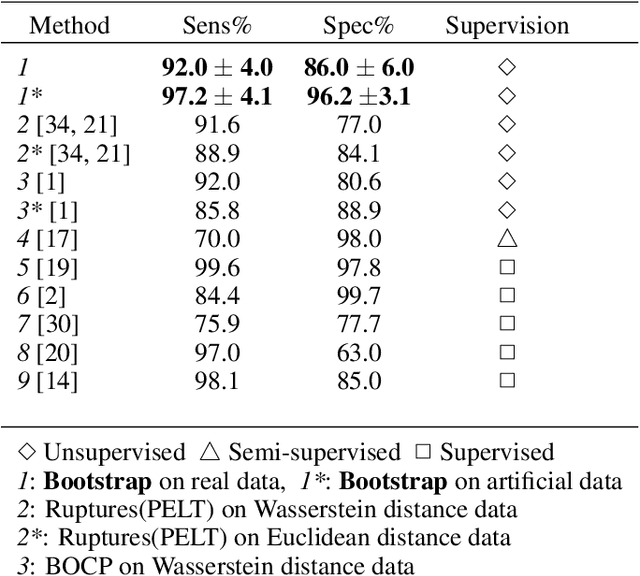
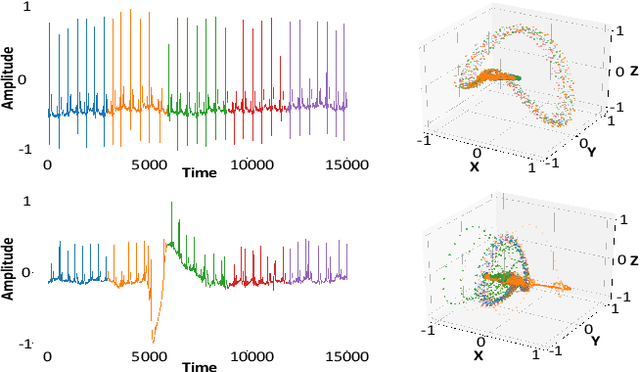
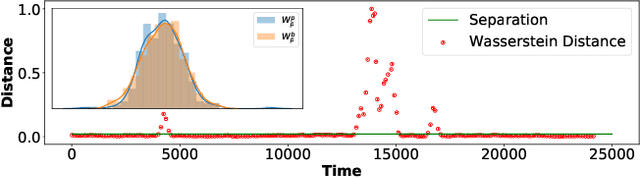
Abstract:We propose a new unsupervised and non-parametric method to detect change points in intricate quasi-periodic signals. The detection relies on optimal transport theory combined with topological analysis and the bootstrap procedure. The algorithm is designed to detect changes in virtually any harmonic or a partially harmonic signal and is verified on three different sources of physiological data streams. We successfully find abnormal or irregular cardiac cycles in the waveforms for the six of the most frequent types of clinical arrhythmias using a single algorithm. The validation and the efficiency of the method are shown both on synthetic and on real time series. Our unsupervised approach reaches the level of performance of the supervised state-of-the-art techniques. We provide conceptual justification for the efficiency of the method and prove the convergence of the bootstrap procedure theoretically.
Microscopy Image Restoration with Deep Wiener-Kolmogorov filters
Dec 25, 2019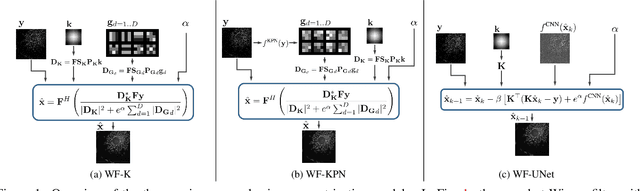
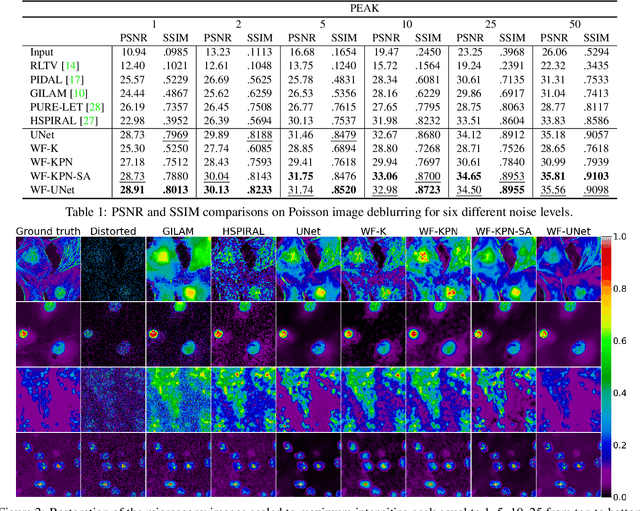
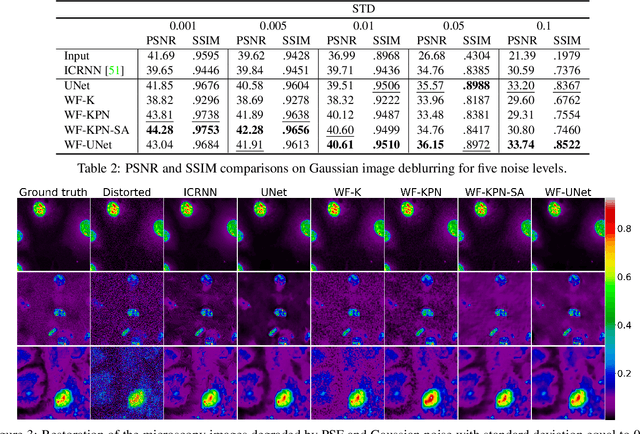
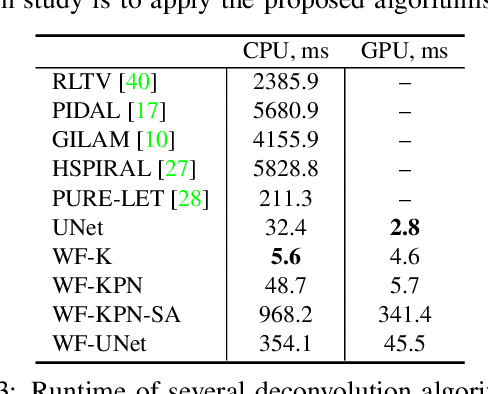
Abstract:Microscopy is a powerful visualization tool in biology, enabling the study of cells, tissues, and the fundamental biological processes. Yet, the observed images of the objects at the micro-scale suffer from two major inherent distortions: the blur caused by the diffraction of light, and the background noise caused by the imperfections of the imaging detectors. The latter is especially severe in fluorescence and in confocal microscopes, which are known for operating at the low photon count with the Poisson noise statistics. Restoration of such images is usually accomplished by image deconvolution, with the nature of the noise statistics taken into account, and by solving an optimization problem given some prior information about the underlying data (i.e., regularization). In this work, we propose a unifying framework of algorithms for Poisson image deblurring and denoising. The algorithms are based on deep learning techniques for the design of learnable regularizers paired with an appropriate optimization scheme. Our extensive experimentation line showcases that the proposed approach achieves superior quality of image reconstruction and beats the solutions that rely on deep learning or on the optimization schemes alone. Moreover, several implementations of the proposed framework demonstrate competitive performance at a low computational complexity, which is of high importance for real-time imaging applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge