Dima Galat
LLM Ensemble for RAG: Role of Context Length in Zero-Shot Question Answering for BioASQ Challenge
Sep 10, 2025



Abstract:Biomedical question answering (QA) poses significant challenges due to the need for precise interpretation of specialized knowledge drawn from a vast, complex, and rapidly evolving corpus. In this work, we explore how large language models (LLMs) can be used for information retrieval (IR), and an ensemble of zero-shot models can accomplish state-of-the-art performance on a domain-specific Yes/No QA task. Evaluating our approach on the BioASQ challenge tasks, we show that ensembles can outperform individual LLMs and in some cases rival or surpass domain-tuned systems - all while preserving generalizability and avoiding the need for costly fine-tuning or labeled data. Our method aggregates outputs from multiple LLM variants, including models from Anthropic and Google, to synthesize more accurate and robust answers. Moreover, our investigation highlights a relationship between context length and performance: while expanded contexts are meant to provide valuable evidence, they simultaneously risk information dilution and model disorientation. These findings emphasize IR as a critical foundation in Retrieval-Augmented Generation (RAG) approaches for biomedical QA systems. Precise, focused retrieval remains essential for ensuring LLMs operate within relevant information boundaries when generating answers from retrieved documents. Our results establish that ensemble-based zero-shot approaches, when paired with effective RAG pipelines, constitute a practical and scalable alternative to domain-tuned systems for biomedical question answering.
Advancing LLM detection in the ALTA 2024 Shared Task: Techniques and Analysis
Dec 26, 2024Abstract:The recent proliferation of AI-generated content has prompted significant interest in developing reliable detection methods. This study explores techniques for identifying AI-generated text through sentence-level evaluation within hybrid articles. Our findings indicate that ChatGPT-3.5 Turbo exhibits distinct, repetitive probability patterns that enable consistent in-domain detection. Empirical tests show that minor textual modifications, such as rewording, have minimal impact on detection accuracy. These results provide valuable insights for advancing AI detection methodologies, offering a pathway toward robust solutions to address the complexities of synthetic text identification.
Enhancing Biomedical Text Summarization and Question-Answering: On the Utility of Domain-Specific Pre-Training
Jul 10, 2023Abstract:Biomedical summarization requires large datasets to train for text generation. We show that while transfer learning offers a viable option for addressing this challenge, an in-domain pre-training does not always offer advantages in a BioASQ summarization task. We identify a suitable model architecture and use it to show a benefit of a general-domain pre-training followed by a task-specific fine-tuning in the context of a BioASQ summarization task, leading to a novel three-step fine-tuning approach that works with only a thousand in-domain examples. Our results indicate that a Large Language Model without domain-specific pre-training can have a significant edge in some domain-specific biomedical text generation tasks.
Query-Focused Extractive Summarisation for Finding Ideal Answers to Biomedical and COVID-19 Questions
Aug 31, 2021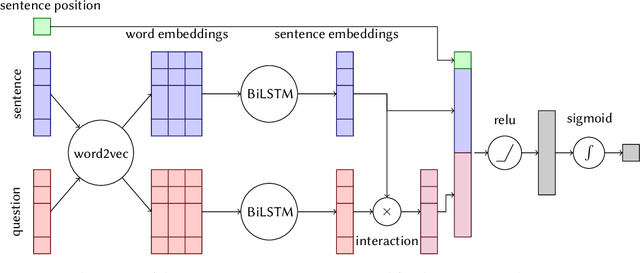
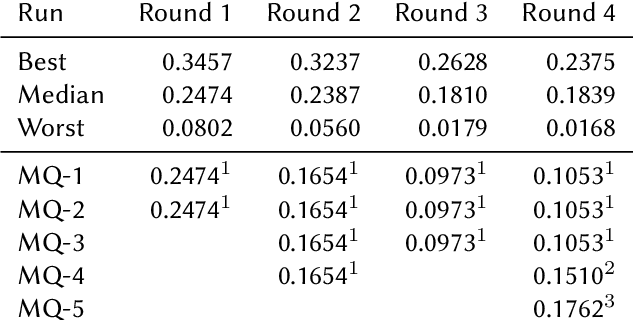
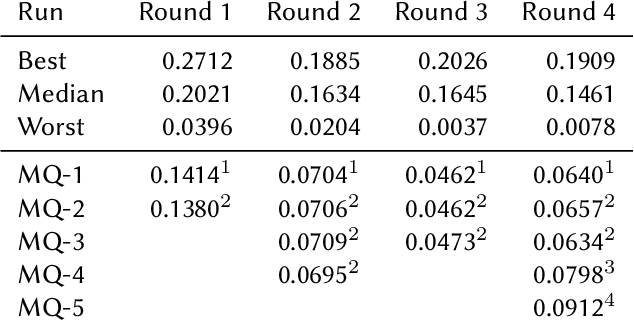
Abstract:This paper presents Macquarie University's participation to the BioASQ Synergy Task, and BioASQ9b Phase B. In each of these tasks, our participation focused on the use of query-focused extractive summarisation to obtain the ideal answers to medical questions. The Synergy Task is an end-to-end question answering task on COVID-19 where systems are required to return relevant documents, snippets, and answers to a given question. Given the absence of training data, we used a query-focused summarisation system that was trained with the BioASQ8b training data set and we experimented with methods to retrieve the documents and snippets. Considering the poor quality of the documents and snippets retrieved by our system, we observed reasonably good quality in the answers returned. For phase B of the BioASQ9b task, the relevant documents and snippets were already included in the test data. Our system split the snippets into candidate sentences and used BERT variants under a sentence classification setup. The system used the question and candidate sentence as input and was trained to predict the likelihood of the candidate sentence being part of the ideal answer. The runs obtained either the best or second best ROUGE-F1 results of all participants to all batches of BioASQ9b. This shows that using BERT in a classification setup is a very strong baseline for the identification of ideal answers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge