Cuong D. Do
PTB-Image: A Scanned Paper ECG Dataset for Digitization and Image-based Diagnosis
Feb 19, 2025Abstract:Electrocardiograms (ECGs) recorded on paper remain prevalent in clinical practice, yet their use presents challenges for automated analysis and digital storage. To address this issue, we introduce PTB-Image, a dataset comprising scanned paper ECGs with corresponding digital signals, enabling research on ECG digitization. We also provide VinDigitizer, a digitization baseline to convert paper-based ECGs into digital time-series signals. The method involves detecting signal rows, extracting waveforms from the background, and reconstructing numerical values from the digitized traces. We applied VinDigitizer to 549 scanned ECGs and evaluated its performance against the original PTB dataset (modified to match the printed signals). The results achieved a mean signal-to-noise ratio (SNR) of 0.01 dB, highlighting both the feasibility and challenges of ECG digitization, particularly in mitigating distortions from printing and scanning processes. By providing PTB-Image and baseline digitization methods, this work aims to facilitate advancements in ECG digitization, enhancing access to historical ECG data and supporting applications in telemedicine and automated cardiac diagnostics.
Transfer Learning in ECG Diagnosis: Is It Effective?
Feb 03, 2024



Abstract:The adoption of deep learning in ECG diagnosis is often hindered by the scarcity of large, well-labeled datasets in real-world scenarios, leading to the use of transfer learning to leverage features learned from larger datasets. Yet the prevailing assumption that transfer learning consistently outperforms training from scratch has never been systematically validated. In this study, we conduct the first extensive empirical study on the effectiveness of transfer learning in multi-label ECG classification, by investigating comparing the fine-tuning performance with that of training from scratch, covering a variety of ECG datasets and deep neural networks. We confirm that fine-tuning is the preferable choice for small downstream datasets; however, when the dataset is sufficiently large, training from scratch can achieve comparable performance, albeit requiring a longer training time to catch up. Furthermore, we find that transfer learning exhibits better compatibility with convolutional neural networks than with recurrent neural networks, which are the two most prevalent architectures for time-series ECG applications. Our results underscore the importance of transfer learning in ECG diagnosis, yet depending on the amount of available data, researchers may opt not to use it, considering the non-negligible cost associated with pre-training.
MPCNN: A Novel Matrix Profile Approach for CNN-based Sleep Apnea Classification
Nov 25, 2023Abstract:Sleep apnea (SA) is a significant respiratory condition that poses a major global health challenge. Previous studies have investigated several machine and deep learning models for electrocardiogram (ECG)-based SA diagnoses. Despite these advancements, conventional feature extractions derived from ECG signals, such as R-peaks and RR intervals, may fail to capture crucial information encompassed within the complete PQRST segments. In this study, we propose an innovative approach to address this diagnostic gap by delving deeper into the comprehensive segments of the ECG signal. The proposed methodology draws inspiration from Matrix Profile algorithms, which generate an Euclidean distance profile from fixed-length signal subsequences. From this, we derived the Min Distance Profile (MinDP), Max Distance Profile (MaxDP), and Mean Distance Profile (MeanDP) based on the minimum, maximum, and mean of the profile distances, respectively. To validate the effectiveness of our approach, we use the modified LeNet-5 architecture as the primary CNN model, along with two existing lightweight models, BAFNet and SE-MSCNN, for ECG classification tasks. Our extensive experimental results on the PhysioNet Apnea-ECG dataset revealed that with the new feature extraction method, we achieved a per-segment accuracy up to 92.11 \% and a per-recording accuracy of 100\%. Moreover, it yielded the highest correlation compared to state-of-the-art methods, with a correlation coefficient of 0.989. By introducing a new feature extraction method based on distance relationships, we enhanced the performance of certain lightweight models, showing potential for home sleep apnea test (HSAT) and SA detection in IoT devices. The source code for this work is made publicly available in GitHub: https://github.com/vinuni-vishc/MPCNN-Sleep-Apnea.
MELEP: A Novel Predictive Measure of Transferability in Multi-Label ECG Analysis
Oct 27, 2023Abstract:We introduce MELEP, which stands for Muti-label Expected Log of Empirical Predictions, a novel measure to estimate how effective it is to transfer knowledge from a pre-trained model to a downstream task in a multi-label settings. The measure is generic to work with new target data having a different label set from source data. It is also computationally efficient, only requires forward passing the downstream dataset through the pre-trained model once. To the best of our knowledge, we are the first to develop such a transferability metric for multi-label ECG classification problems. Our experiments show that MELEP can predict the performance of pre-trained convolutional and recurrent deep neural networks, on small and imbalanced ECG data. Specifically, strong correlation coefficients, with absolute values exceeding 0.6 in most cases, were observed between MELEP and the actual average F1 scores of the fine-tuned models.
Enhancing Deep Learning-based 3-lead ECG Classification with Heartbeat Counting and Demographic Data Integration
Aug 15, 2022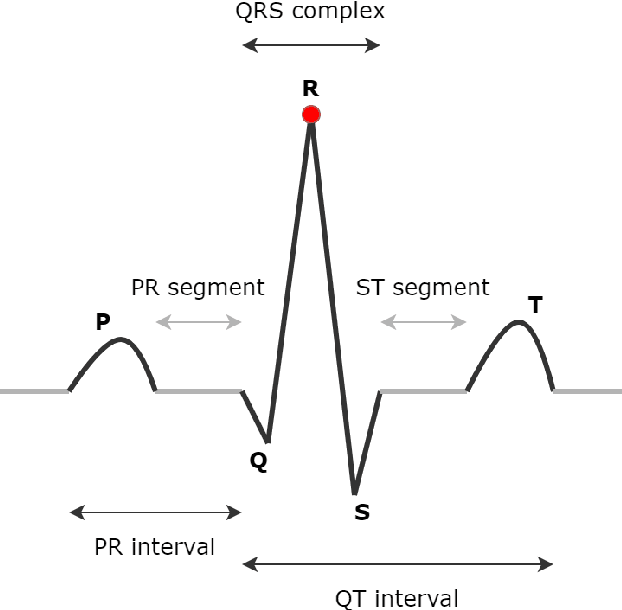
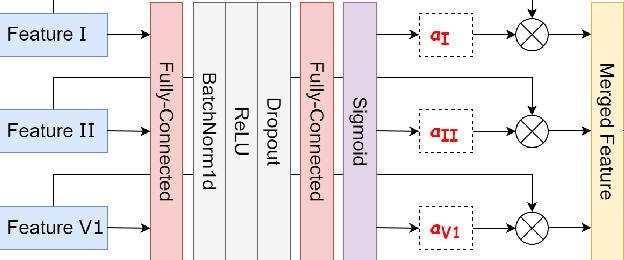
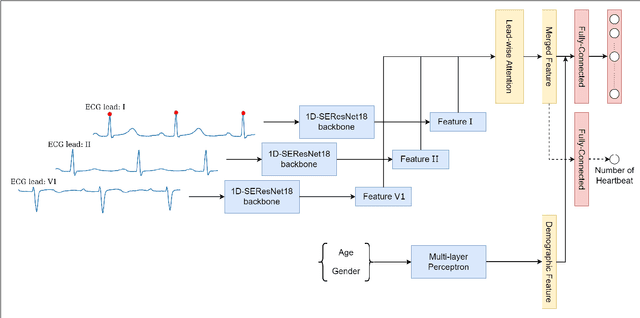
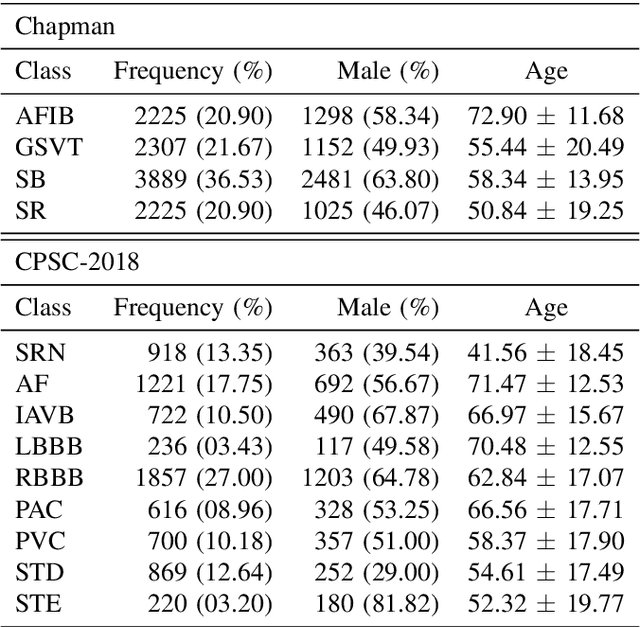
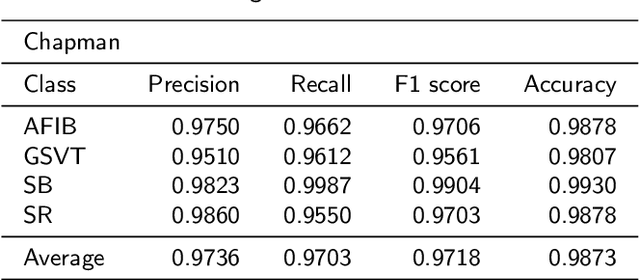
Abstract:Nowadays, an increasing number of people are being diagnosed with cardiovascular diseases (CVDs), the leading cause of death globally. The gold standard for identifying these heart problems is via electrocardiogram (ECG). The standard 12-lead ECG is widely used in clinical practice and the majority of current research. However, using a lower number of leads can make ECG more pervasive as it can be integrated with portable or wearable devices. This article introduces two novel techniques to improve the performance of the current deep learning system for 3-lead ECG classification, making it comparable with models that are trained using standard 12-lead ECG. Specifically, we propose a multi-task learning scheme in the form of the number of heartbeats regression and an effective mechanism to integrate patient demographic data into the system. With these two advancements, we got classification performance in terms of F1 scores of 0.9796 and 0.8140 on two large-scale ECG datasets, i.e., Chapman and CPSC-2018, respectively, which surpassed current state-of-the-art ECG classification methods, even those trained on 12-lead data. To encourage further development, our source code is publicly available at https://github.com/lhkhiem28/LightX3ECG.
LightX3ECG: A Lightweight and eXplainable Deep Learning System for 3-lead Electrocardiogram Classification
Jul 25, 2022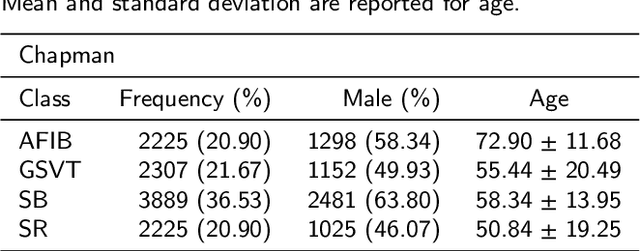
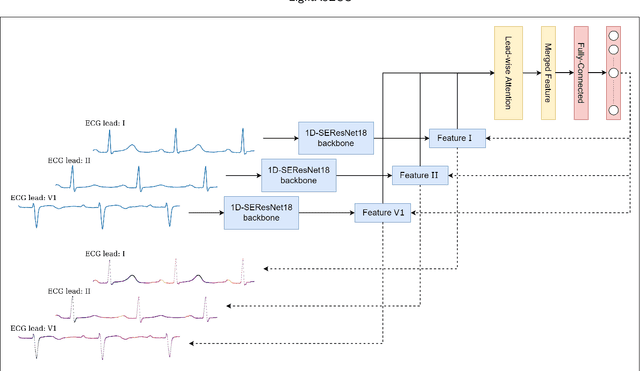
Abstract:Cardiovascular diseases (CVDs) are a group of heart and blood vessel disorders that is one of the most serious dangers to human health, and the number of such patients is still growing. Early and accurate detection plays a key role in successful treatment and intervention. Electrocardiogram (ECG) is the gold standard for identifying a variety of cardiovascular abnormalities. In clinical practices and most of the current research, standard 12-lead ECG is mainly used. However, using a lower number of leads can make ECG more prevalent as it can be conveniently recorded by portable or wearable devices. In this research, we develop a novel deep learning system to accurately identify multiple cardiovascular abnormalities by using only three ECG leads.
VinDr-CXR: An open dataset of chest X-rays with radiologist's annotations
Jan 03, 2021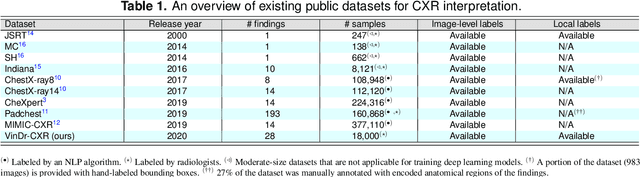
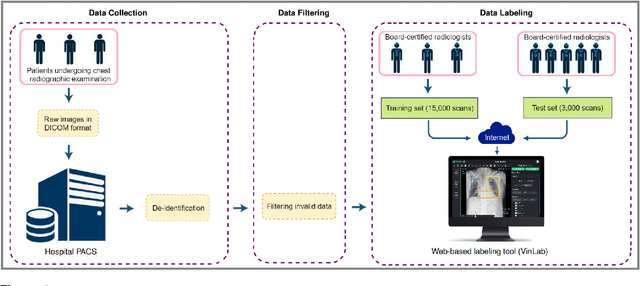
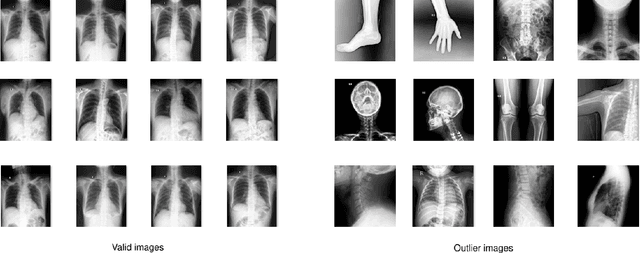
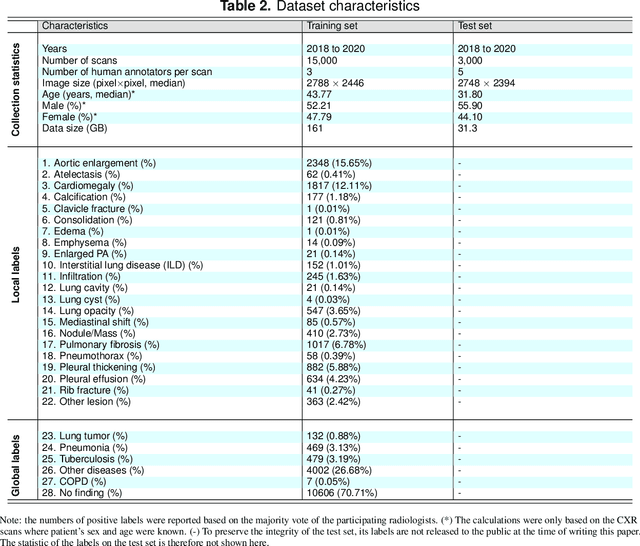
Abstract:Most of the existing chest X-ray datasets include labels from a list of findings without specifying their locations on the radiographs. This limits the development of machine learning algorithms for the detection and localization of chest abnormalities. In this work, we describe a dataset of more than 100,000 chest X-ray scans that were retrospectively collected from two major hospitals in Vietnam. Out of this raw data, we release 18,000 images that were manually annotated by a total of 17 experienced radiologists with 22 local labels of rectangles surrounding abnormalities and 6 global labels of suspected diseases. The released dataset is divided into a training set of 15,000 and a test set of 3,000. Each scan in the training set was independently labeled by 3 radiologists, while each scan in the test set was labeled by the consensus of 5 radiologists. We designed and built a labeling platform for DICOM images to facilitate these annotation procedures. All images are made publicly available in DICOM format in company with the labels of the training set. The labels of the test set are hidden at the time of writing this paper as they will be used for benchmarking machine learning algorithms on an open platform.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge