Linh T. Le
Evaluating the impact of an explainable machine learning system on the interobserver agreement in chest radiograph interpretation
Apr 01, 2023

Abstract:We conducted a prospective study to measure the clinical impact of an explainable machine learning system on interobserver agreement in chest radiograph interpretation. The AI system, which we call as it VinDr-CXR when used as a diagnosis-supporting tool, significantly improved the agreement between six radiologists with an increase of 1.5% in mean Fleiss' Kappa. In addition, we also observed that, after the radiologists consulted AI's suggestions, the agreement between each radiologist and the system was remarkably increased by 3.3% in mean Cohen's Kappa. This work has been accepted for publication in IEEE Access and this paper is our short version submitted to the Midwest Machine Learning Symposium (MMLS 2023), Chicago, IL, USA.
An Accurate and Explainable Deep Learning System Improves Interobserver Agreement in the Interpretation of Chest Radiograph
Aug 06, 2022Abstract:Recent artificial intelligence (AI) algorithms have achieved radiologist-level performance on various medical classification tasks. However, only a few studies addressed the localization of abnormal findings from CXR scans, which is essential in explaining the image-level classification to radiologists. We introduce in this paper an explainable deep learning system called VinDr-CXR that can classify a CXR scan into multiple thoracic diseases and, at the same time, localize most types of critical findings on the image. VinDr-CXR was trained on 51,485 CXR scans with radiologist-provided bounding box annotations. It demonstrated a comparable performance to experienced radiologists in classifying 6 common thoracic diseases on a retrospective validation set of 3,000 CXR scans, with a mean area under the receiver operating characteristic curve (AUROC) of 0.967 (95% confidence interval [CI]: 0.958-0.975). The VinDr-CXR was also externally validated in independent patient cohorts and showed its robustness. For the localization task with 14 types of lesions, our free-response receiver operating characteristic (FROC) analysis showed that the VinDr-CXR achieved a sensitivity of 80.2% at the rate of 1.0 false-positive lesion identified per scan. A prospective study was also conducted to measure the clinical impact of the VinDr-CXR in assisting six experienced radiologists. The results indicated that the proposed system, when used as a diagnosis supporting tool, significantly improved the agreement between radiologists themselves with an increase of 1.5% in mean Fleiss' Kappa. We also observed that, after the radiologists consulted VinDr-CXR's suggestions, the agreement between each of them and the system was remarkably increased by 3.3% in mean Cohen's Kappa.
VinDr-Mammo: A large-scale benchmark dataset for computer-aided diagnosis in full-field digital mammography
Mar 20, 2022Abstract:Mammography, or breast X-ray, is the most widely used imaging modality to detect cancer and other breast diseases. Recent studies have shown that deep learning-based computer-assisted detection and diagnosis (CADe or CADx) tools have been developed to support physicians and improve the accuracy of interpreting mammography. However, most published datasets of mammography are either limited on sample size or digitalized from screen-film mammography (SFM), hindering the development of CADe and CADx tools which are developed based on full-field digital mammography (FFDM). To overcome this challenge, we introduce VinDr-Mammo - a new benchmark dataset of FFDM for detecting and diagnosing breast cancer and other diseases in mammography. The dataset consists of 5,000 mammography exams, each of which has four standard views and is double read with disagreement (if any) being resolved by arbitration. It is created for the assessment of Breast Imaging Reporting and Data System (BI-RADS) and density at the breast level. In addition, the dataset also provides the category, location, and BI-RADS assessment of non-benign findings. We make VinDr-Mammo publicly available on PhysioNet as a new imaging resource to promote advances in developing CADe and CADx tools for breast cancer screening.
VinDr-CXR: An open dataset of chest X-rays with radiologist's annotations
Jan 03, 2021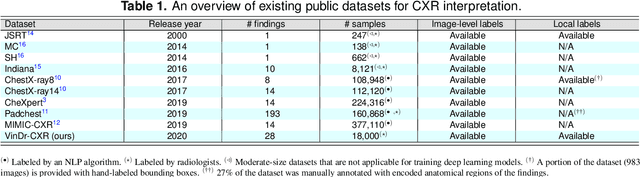
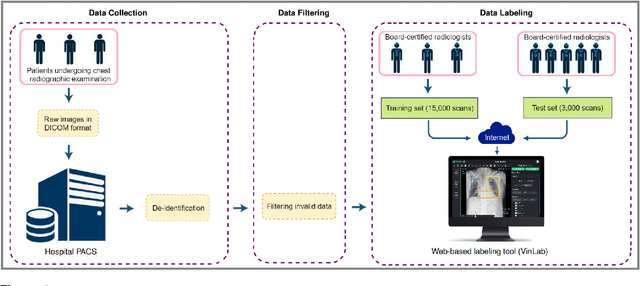
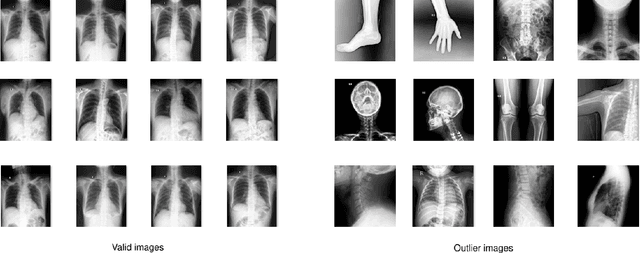
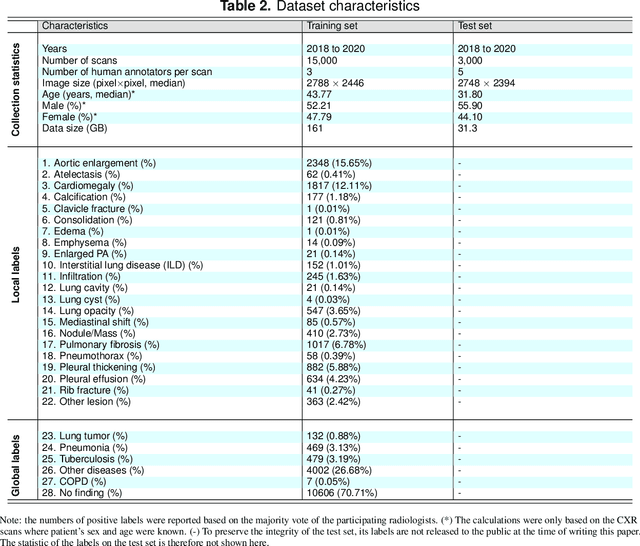
Abstract:Most of the existing chest X-ray datasets include labels from a list of findings without specifying their locations on the radiographs. This limits the development of machine learning algorithms for the detection and localization of chest abnormalities. In this work, we describe a dataset of more than 100,000 chest X-ray scans that were retrospectively collected from two major hospitals in Vietnam. Out of this raw data, we release 18,000 images that were manually annotated by a total of 17 experienced radiologists with 22 local labels of rectangles surrounding abnormalities and 6 global labels of suspected diseases. The released dataset is divided into a training set of 15,000 and a test set of 3,000. Each scan in the training set was independently labeled by 3 radiologists, while each scan in the test set was labeled by the consensus of 5 radiologists. We designed and built a labeling platform for DICOM images to facilitate these annotation procedures. All images are made publicly available in DICOM format in company with the labels of the training set. The labels of the test set are hidden at the time of writing this paper as they will be used for benchmarking machine learning algorithms on an open platform.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge