Claudio Santiago
Deep Symbolic Optimization: Reinforcement Learning for Symbolic Mathematics
May 16, 2025Abstract:Deep Symbolic Optimization (DSO) is a novel computational framework that enables symbolic optimization for scientific discovery, particularly in applications involving the search for intricate symbolic structures. One notable example is equation discovery, which aims to automatically derive mathematical models expressed in symbolic form. In DSO, the discovery process is formulated as a sequential decision-making task. A generative neural network learns a probabilistic model over a vast space of candidate symbolic expressions, while reinforcement learning strategies guide the search toward the most promising regions. This approach integrates gradient-based optimization with evolutionary and local search techniques, and it incorporates in-situ constraints, domain-specific priors, and advanced policy optimization methods. The result is a robust framework capable of efficiently exploring extensive search spaces to identify interpretable and physically meaningful models. Extensive evaluations on benchmark problems have demonstrated that DSO achieves state-of-the-art performance in both accuracy and interpretability. In this chapter, we provide a comprehensive overview of the DSO framework and illustrate its transformative potential for automating symbolic optimization in scientific discovery.
Precision medicine as a control problem: Using simulation and deep reinforcement learning to discover adaptive, personalized multi-cytokine therapy for sepsis
Feb 08, 2018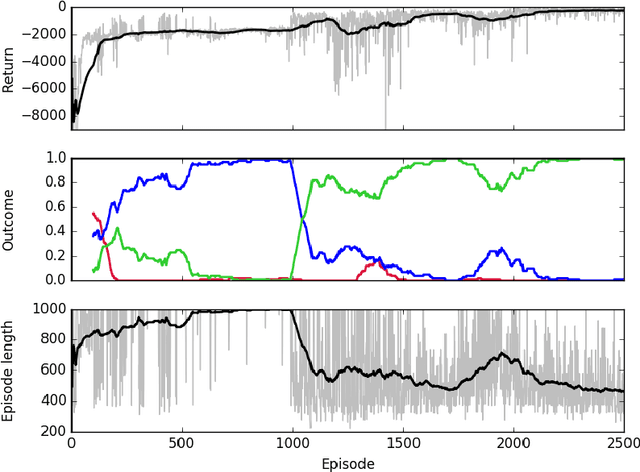
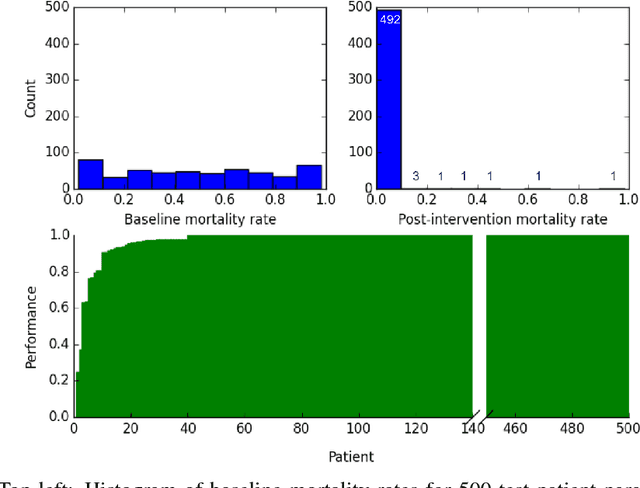
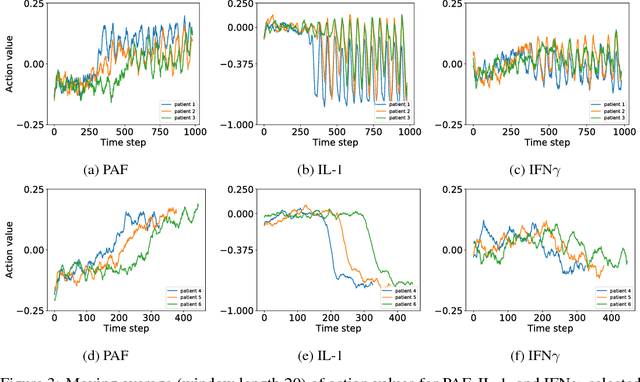
Abstract:Sepsis is a life-threatening condition affecting one million people per year in the US in which dysregulation of the body's own immune system causes damage to its tissues, resulting in a 28 - 50% mortality rate. Clinical trials for sepsis treatment over the last 20 years have failed to produce a single currently FDA approved drug treatment. In this study, we attempt to discover an effective cytokine mediation treatment strategy for sepsis using a previously developed agent-based model that simulates the innate immune response to infection: the Innate Immune Response agent-based model (IIRABM). Previous attempts at reducing mortality with multi-cytokine mediation using the IIRABM have failed to reduce mortality across all patient parameterizations and motivated us to investigate whether adaptive, personalized multi-cytokine mediation can control the trajectory of sepsis and lower patient mortality. We used the IIRABM to compute a treatment policy in which systemic patient measurements are used in a feedback loop to inform future treatment. Using deep reinforcement learning, we identified a policy that achieves 0% mortality on the patient parameterization on which it was trained. More importantly, this policy also achieves 0.8% mortality over 500 randomly selected patient parameterizations with baseline mortalities ranging from 1 - 99% (with an average of 49%) spanning the entire clinically plausible parameter space of the IIRABM. These results suggest that adaptive, personalized multi-cytokine mediation therapy could be a promising approach for treating sepsis. We hope that this work motivates researchers to consider such an approach as part of future clinical trials. To the best of our knowledge, this work is the first to consider adaptive, personalized multi-cytokine mediation therapy for sepsis, and is the first to exploit deep reinforcement learning on a biological simulation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge