Christian Rummel
Exploring Robustness of Cortical Morphometry in the presence of white matter lesions, using Diffusion Models for Lesion Filling
Mar 26, 2025Abstract:Cortical thickness measurements from magnetic resonance imaging, an important biomarker in many neurodegenerative and neurological disorders, are derived by many tools from an initial voxel-wise tissue segmentation. White matter (WM) hypointensities in T1-weighted imaging, such as those arising from multiple sclerosis or small vessel disease, are known to affect the output of brain segmentation methods and therefore bias cortical thickness measurements. These effects are well-documented among traditional brain segmentation tools but have not been studied extensively in tools based on deep-learning segmentations, which promise to be more robust. In this paper, we explore the potential of deep learning to enhance the accuracy and efficiency of cortical thickness measurement in the presence of WM lesions, using a high-quality lesion filling algorithm leveraging denoising diffusion networks. A pseudo-3D U-Net architecture trained on the OASIS dataset to generate synthetic healthy tissue, conditioned on binary lesion masks derived from the MSSEG dataset, allows realistic removal of white matter lesions in multiple sclerosis patients. By applying morphometry methods to patient images before and after lesion filling, we analysed robustness of global and regional cortical thickness measurements in the presence of white matter lesions. Methods based on a deep learning-based segmentation of the brain (Fastsurfer, DL+DiReCT, ANTsPyNet) exhibited greater robustness than those using classical segmentation methods (Freesurfer, ANTs).
CortexMorph: fast cortical thickness estimation via diffeomorphic registration using VoxelMorph
Jul 21, 2023



Abstract:The thickness of the cortical band is linked to various neurological and psychiatric conditions, and is often estimated through surface-based methods such as Freesurfer in MRI studies. The DiReCT method, which calculates cortical thickness using a diffeomorphic deformation of the gray-white matter interface towards the pial surface, offers an alternative to surface-based methods. Recent studies using a synthetic cortical thickness phantom have demonstrated that the combination of DiReCT and deep-learning-based segmentation is more sensitive to subvoxel cortical thinning than Freesurfer. While anatomical segmentation of a T1-weighted image now takes seconds, existing implementations of DiReCT rely on iterative image registration methods which can take up to an hour per volume. On the other hand, learning-based deformable image registration methods like VoxelMorph have been shown to be faster than classical methods while improving registration accuracy. This paper proposes CortexMorph, a new method that employs unsupervised deep learning to directly regress the deformation field needed for DiReCT. By combining CortexMorph with a deep-learning-based segmentation model, it is possible to estimate region-wise thickness in seconds from a T1-weighted image, while maintaining the ability to detect cortical atrophy. We validate this claim on the OASIS-3 dataset and the synthetic cortical thickness phantom of Rusak et al.
Automatic detection of lesion load change in Multiple Sclerosis using convolutional neural networks with segmentation confidence
Apr 05, 2019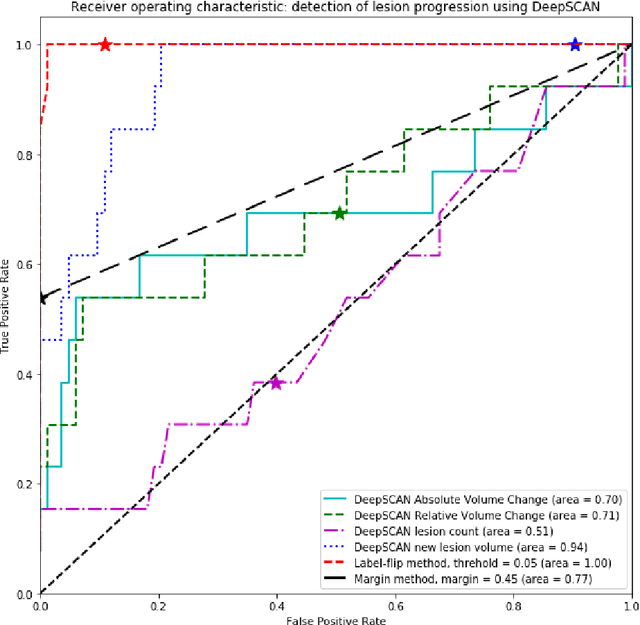
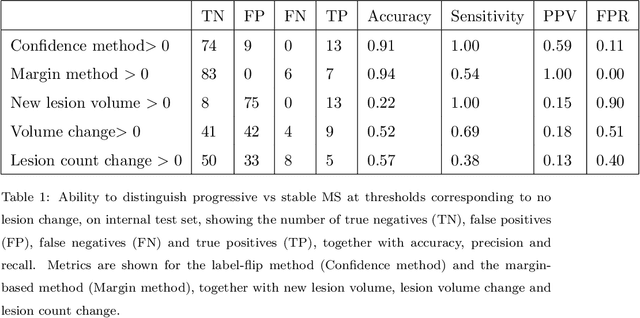
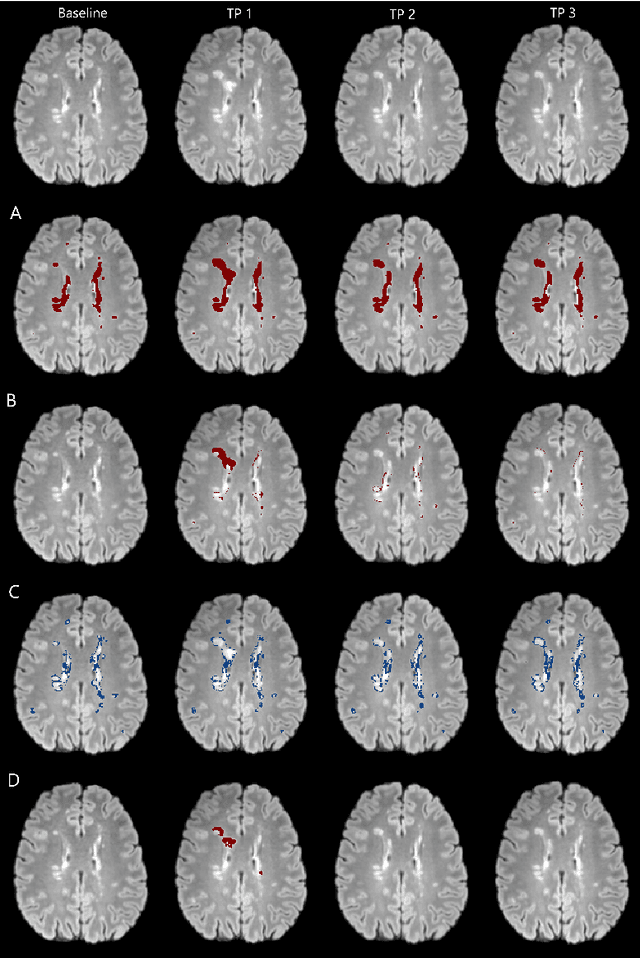
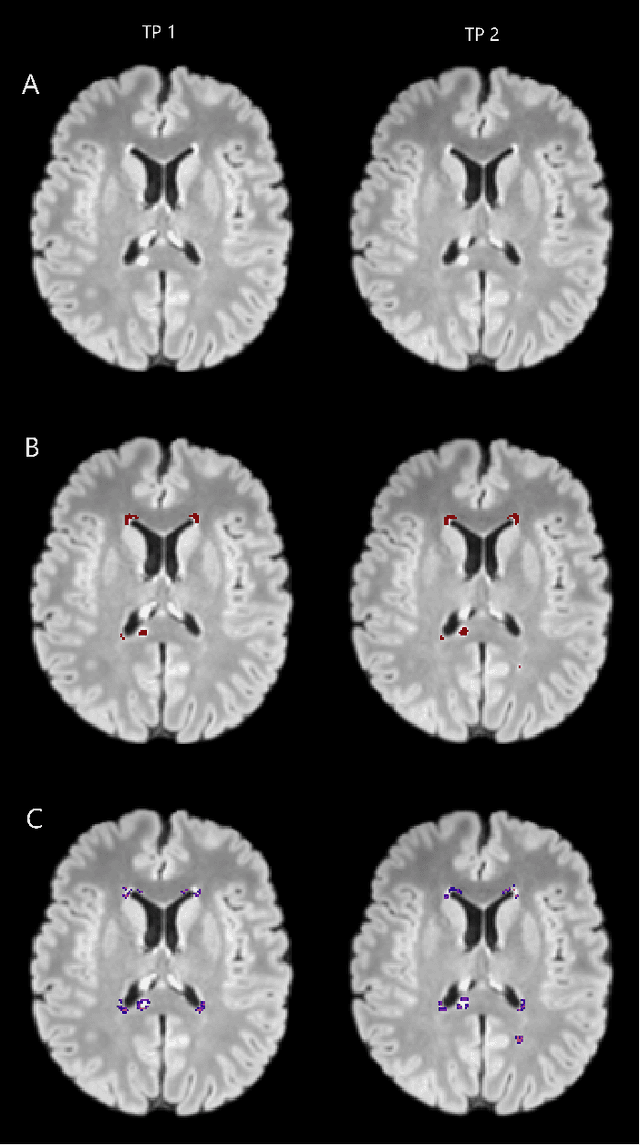
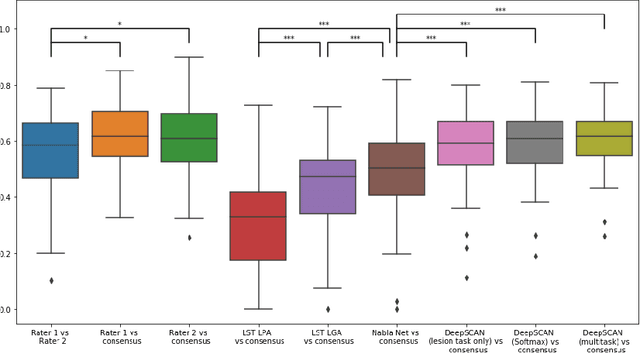
Abstract:The detection of new or enlarged white-matter lesions in multiple sclerosis is a vital task in the monitoring of patients undergoing disease-modifying treatment for multiple sclerosis. However, the definition of 'new or enlarged' is not fixed, and it is known that lesion-counting is highly subjective, with high degree of inter- and intra-rater variability. Automated methods for lesion quantification hold the potential to make the detection of new and enlarged lesions consistent and repeatable. However, the majority of lesion segmentation algorithms are not evaluated for their ability to separate progressive from stable patients, despite this being a pressing clinical use-case. In this paper we show that change in volumetric measurements of lesion load alone is not a good method for performing this separation, even for highly performing segmentation methods. Instead, we propose a method for identifying lesion changes of high certainty, and establish on a dataset of longitudinal multiple sclerosis cases that this method is able to separate progressive from stable timepoints with a very high level of discrimination (AUC = 0.99), while changes in lesion volume are much less able to perform this separation (AUC = 0.71). Validation of the method on a second external dataset confirms that the method is able to generalize beyond the setting in which it was trained, achieving an accuracy of 83% in separating stable and progressive timepoints. Both lesion volume and count have previously been shown to be strong predictors of disease course across a population. However, we demonstrate that for individual patients, changes in these measures are not an adequate means of establishing no evidence of disease activity. Meanwhile, directly detecting tissue which changes, with high confidence, from non-lesion to lesion is a feasible methodology for identifying radiologically active patients.
Few-shot brain segmentation from weakly labeled data with deep heteroscedastic multi-task networks
Apr 04, 2019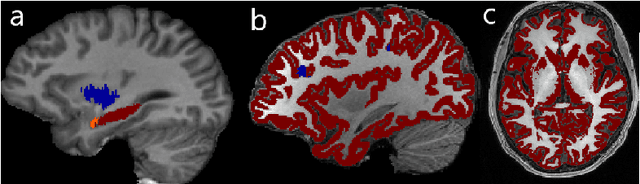
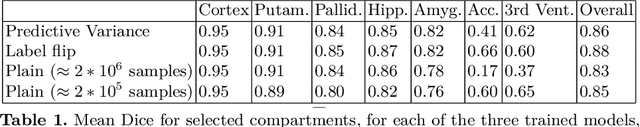
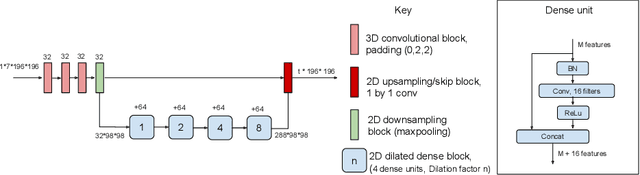
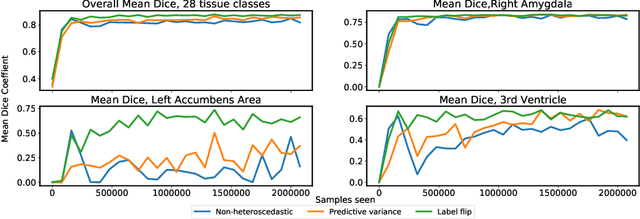
Abstract:In applications of supervised learning applied to medical image segmentation, the need for large amounts of labeled data typically goes unquestioned. In particular, in the case of brain anatomy segmentation, hundreds or thousands of weakly-labeled volumes are often used as training data. In this paper, we first observe that for many brain structures, a small number of training examples, (n=9), weakly labeled using Freesurfer 6.0, plus simple data augmentation, suffice as training data to achieve high performance, achieving an overall mean Dice coefficient of $0.84 \pm 0.12$ compared to Freesurfer over 28 brain structures in T1-weighted images of $\approx 4000$ 9-10 year-olds from the Adolescent Brain Cognitive Development study. We then examine two varieties of heteroscedastic network as a method for improving classification results. An existing proposal by Kendall and Gal, which uses Monte-Carlo inference to learn to predict the variance of each prediction, yields an overall mean Dice of $0.85 \pm 0.14$ and showed statistically significant improvements over 25 brain structures. Meanwhile a novel heteroscedastic network which directly learns the probability that an example has been mislabeled yielded an overall mean Dice of $0.87 \pm 0.11$ and showed statistically significant improvements over all but one of the brain structures considered. The loss function associated to this network can be interpreted as performing a form of learned label smoothing, where labels are only smoothed where they are judged to be uncertain.
Simultaneous lesion and neuroanatomy segmentation in Multiple Sclerosis using deep neural networks
Jan 22, 2019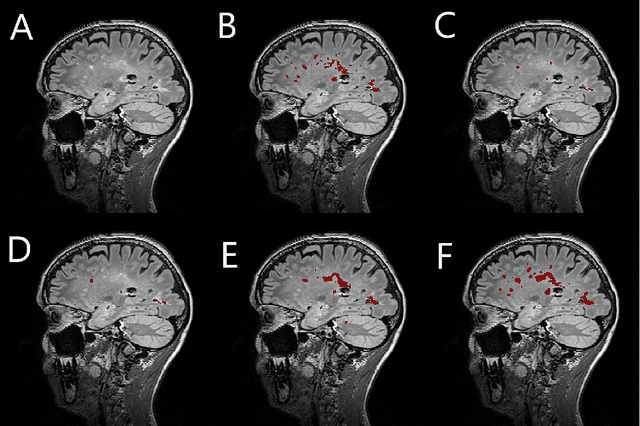
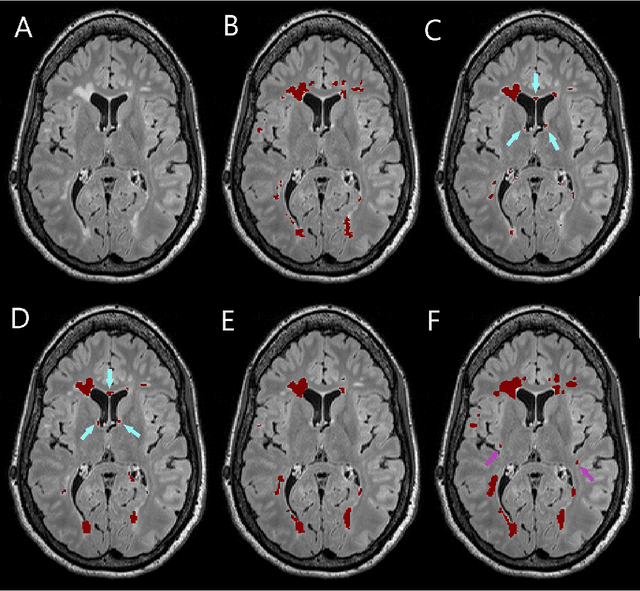
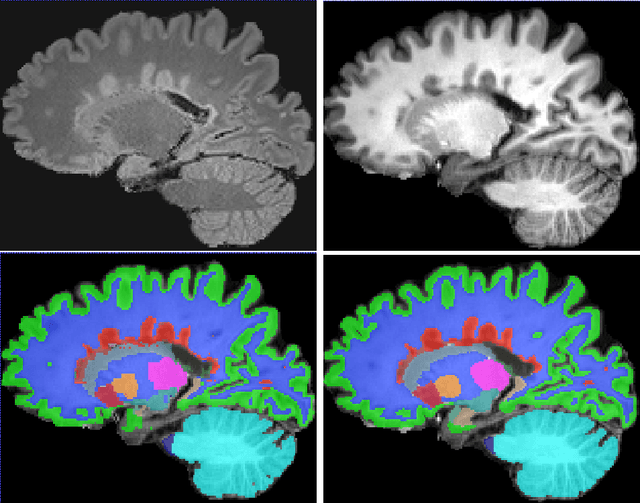
Abstract:Segmentation of both white matter lesions and deep grey matter structures is an important task in the quantification of magnetic resonance imaging in multiple sclerosis. Typically these tasks are performed separately: in this paper we present a single CNN-based segmentation solution for providing fast, reliable segmentations of multimodal MR imagies into lesion classes and healthy-appearing grey- and white-matter structures. We show substantial, statistically significant improvements in both Dice coefficient and in lesion-wise specificity and sensitivity, compared to previous approaches, and agreement with individual human raters in the range of human inter-rater variability. The method is trained on data gathered from a single centre: nonetheless, it performs well on data from centres, scanners and field-strengths not represented in the training dataset. A retrospective study found that the classifier successfully identified lesions missed by the human raters. Lesion labels were provided by human raters, while weak labels for other brain structures (including CSF, cortical grey matter, cortical white matter, cerebellum, amygdala, hippocampus, subcortical GM structures and choroid plexus) were provided by Freesurfer 5.3. The segmentations of these structures compared well, not only with Freesurfer 5.3, but also with FSL-First and Freesurfer 6.1.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge