Chenhao Guo
An Out-Of-Distribution Membership Inference Attack Approach for Cross-Domain Graph Attacks
May 26, 2025Abstract:Graph Neural Network-based methods face privacy leakage risks due to the introduction of topological structures about the targets, which allows attackers to bypass the target's prior knowledge of the sensitive attributes and realize membership inference attacks (MIA) by observing and analyzing the topology distribution. As privacy concerns grow, the assumption of MIA, which presumes that attackers can obtain an auxiliary dataset with the same distribution, is increasingly deviating from reality. In this paper, we categorize the distribution diversity issue in real-world MIA scenarios as an Out-Of-Distribution (OOD) problem, and propose a novel Graph OOD Membership Inference Attack (GOOD-MIA) to achieve cross-domain graph attacks. Specifically, we construct shadow subgraphs with distributions from different domains to model the diversity of real-world data. We then explore the stable node representations that remain unchanged under external influences and consider eliminating redundant information from confounding environments and extracting task-relevant key information to more clearly distinguish between the characteristics of training data and unseen data. This OOD-based design makes cross-domain graph attacks possible. Finally, we perform risk extrapolation to optimize the attack's domain adaptability during attack inference to generalize the attack to other domains. Experimental results demonstrate that GOOD-MIA achieves superior attack performance in datasets designed for multiple domains.
Source-Aware Spatial-Spectral-Integrated Double U-Net for Image Fusion
Dec 13, 2022Abstract:In image fusion tasks, pictures from different sources possess distinctive properties, therefore treating them equally will lead to inadequate feature extracting. Besides, multi-scaled networks capture information more sufficiently than single-scaled models in pixel-wised problems. In light of these factors, we propose a source-aware spatial-spectral-integrated double U-shaped network called $\rm{(SU)^2}$Net. The network is mainly composed of a spatial U-net and a spectral U-net, which learn spatial details and spectral characteristics discriminately and hierarchically. In contrast with most previous works that simply apply concatenation to integrate spatial and spectral information, a novel structure named the spatial-spectral block (called $\rm{S^2}$Block) is specially designed to merge feature maps from different sources effectively. Experiment results show that our method outperforms the representative state-of-the-art (SOTA) approaches in both quantitative and qualitative evaluations for a variety of image fusion missions, including remote sensing pansharpening and hyperspectral image super-resolution (HISR).
Wasserstein Adversarial Autoencoders for Knowledge Graph Embedding based Drug-Drug Interaction Prediction
Apr 15, 2020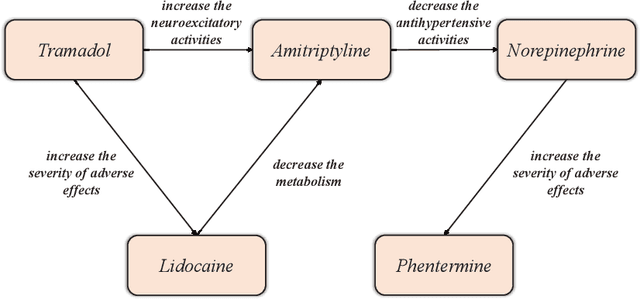

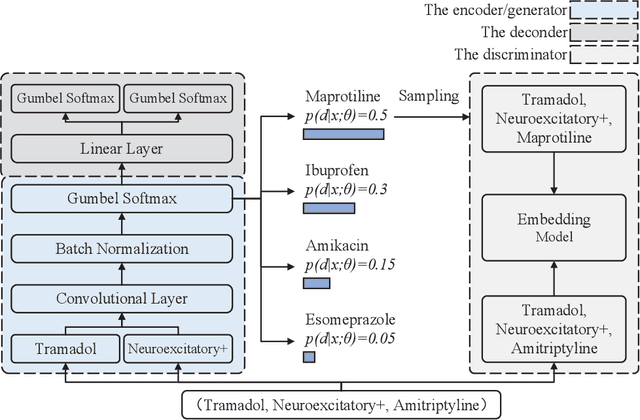
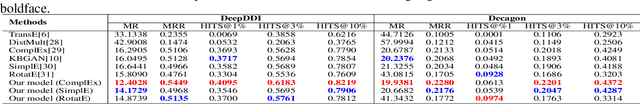
Abstract:Interaction between pharmacological agents can trigger unexpected adverse events. Capturing richer and more comprehensive information about drug-drug interactions (DDI) is one of the key tasks in public health and drug development. Recently, several knowledge graph embedding approaches have received increasing attention in the DDI domain due to their capability of projecting drugs and interactions into a low-dimensional feature space for predicting links and classifying triplets. However, existing methods only apply a uniformly random mode to construct negative samples. As a consequence, these samples are often too simplistic to train an effective model. In this paper, we propose a new knowledge graph embedding framework by introducing adversarial autoencoders (AAE) based on Wasserstein distances and Gumbel-Softmax relaxation for drug-drug interactions tasks. In our framework, the autoencoder is employed to generate high-quality negative samples and the hidden vector of the autoencoder is regarded as a plausible drug candidate. Afterwards, the discriminator learns the embeddings of drugs and interactions based on both positive and negative triplets. Meanwhile, in order to solve vanishing gradient problems on the discrete representation--an inherent flaw in traditional generative models--we utilize the Gumbel-Softmax relaxation and the Wasserstein distance to train the embedding model steadily. We empirically evaluate our method on two tasks, link prediction and DDI classification. The experimental results show that our framework can attain significant improvements and noticeably outperform competitive baselines.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge