Chen-Han Tsai
Revisiting Data Compression with Language Modeling
Jan 06, 2026Abstract:In this report, we investigate the potential use of large language models (LLM's) in the task of data compression. Previous works have demonstrated promising results in applying LLM's towards compressing not only text, but also a wide range of multi-modal data. Despite the favorable performance achieved, there still remains several practical questions that pose a challenge towards replacing existing data compression algorithms with LLM's. In this work, we explore different methods to achieve a lower adjusted compression rate using LLM's as data compressors. In comparison to previous works, we were able to achieve a new state-of-the-art (SOTA) adjusted compression rate of around $18\%$ on the enwik9 dataset without additional model training. Furthermore, we explore the use of LLM's in compressing non-English data, code data, byte stream sequences. We show that while LLM's excel in compressing data in text-dominant domains, their ability in compressing non-natural text sequences still remain competitive if configured in the right way.
DeCode: Decoupling Content and Delivery for Medical QA
Jan 05, 2026Abstract:Large language models (LLMs) exhibit strong medical knowledge and can generate factually accurate responses. However, existing models often fail to account for individual patient contexts, producing answers that are clinically correct yet poorly aligned with patients' needs. In this work, we introduce DeCode, a training-free, model-agnostic framework that adapts existing LLMs to produce contextualized answers in clinical settings. We evaluate DeCode on OpenAI HealthBench, a comprehensive and challenging benchmark designed to assess clinical relevance and validity of LLM responses. DeCode improves the previous state of the art from $28.4\%$ to $49.8\%$, corresponding to a $75\%$ relative improvement. Experimental results suggest the effectiveness of DeCode in improving clinical question answering of LLMs.
RANSAC-NN: Unsupervised Image Outlier Detection using RANSAC
Jul 23, 2023Abstract:Image outlier detection (OD) is crucial for ensuring the quality and accuracy of image datasets used in computer vision tasks. The majority of OD algorithms, however, have not been targeted toward image data. Consequently, the results of applying such algorithms to images are often suboptimal. In this work, we propose RANSAC-NN, a novel unsupervised OD algorithm specifically designed for images. By comparing images in a RANSAC-based approach, our algorithm automatically predicts the outlier score of each image without additional training or label information. We evaluate RANSAC-NN against state-of-the-art OD algorithms on 15 diverse datasets. Without any hyperparameter tuning, RANSAC-NN consistently performs favorably in contrast to other algorithms in almost every dataset category. Furthermore, we provide a detailed analysis to understand each RANSAC-NN component, and we demonstrate its potential applications in image mislabeled detection. Code for RANSAC-NN is provided at https://github.com/mxtsai/ransac-nn
Multi-Task Lung Nodule Detection in Chest Radiographs with a Dual Head Network
Jul 07, 2022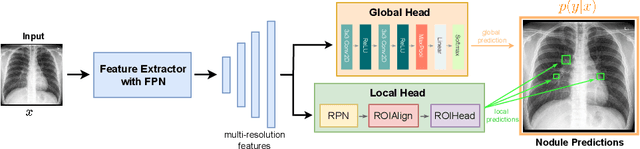
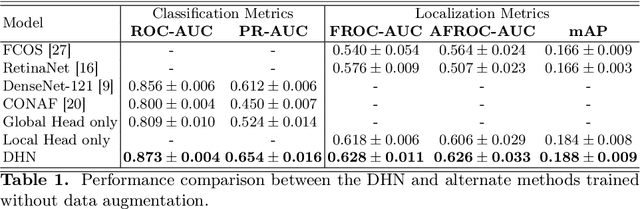
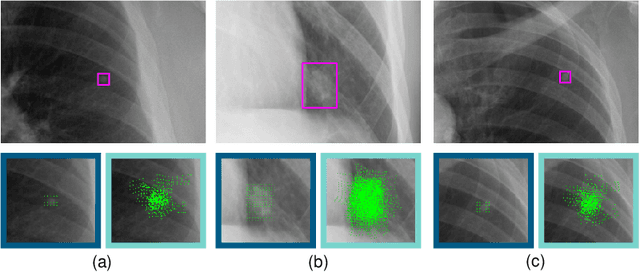
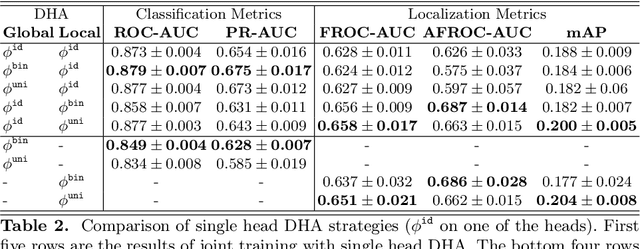
Abstract:Lung nodules can be an alarming precursor to potential lung cancer. Missed nodule detections during chest radiograph analysis remains a common challenge among thoracic radiologists. In this work, we present a multi-task lung nodule detection algorithm for chest radiograph analysis. Unlike past approaches, our algorithm predicts a global-level label indicating nodule presence along with local-level labels predicting nodule locations using a Dual Head Network (DHN). We demonstrate the favorable nodule detection performance that our multi-task formulation yields in comparison to conventional methods. In addition, we introduce a novel Dual Head Augmentation (DHA) strategy tailored for DHN, and we demonstrate its significance in further enhancing global and local nodule predictions.
Labeling of Multilingual Breast MRI Reports
Jul 06, 2020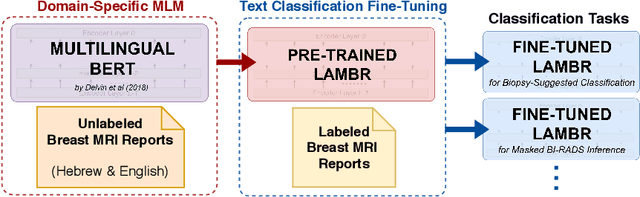
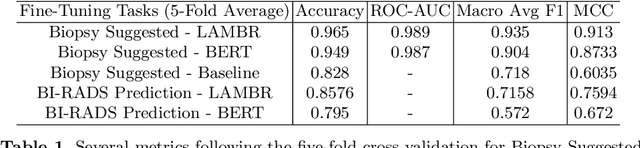
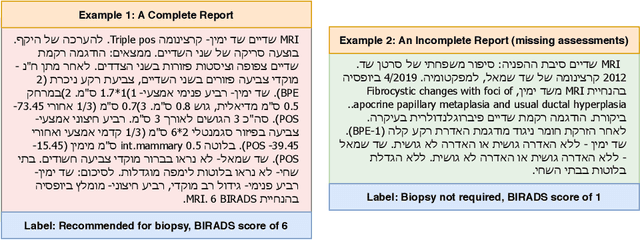
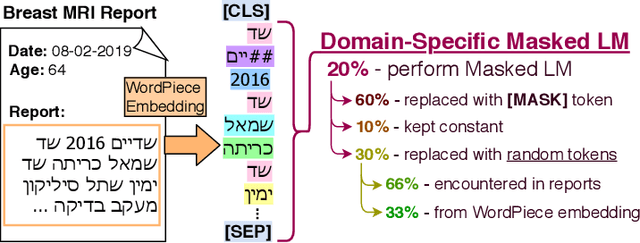
Abstract:Medical reports are an essential medium in recording a patient's condition throughout a clinical trial. They contain valuable information that can be extracted to generate a large labeled dataset needed for the development of clinical tools. However, the majority of medical reports are stored in an unregularized format, and a trained human annotator (typically a doctor) must manually assess and label each case, resulting in an expensive and time consuming procedure. In this work, we present a framework for developing a multilingual breast MRI report classifier using a custom-built language representation called LAMBR. Our proposed method overcomes practical challenges faced in clinical settings, and we demonstrate improved performance in extracting labels from medical reports when compared with conventional approaches.
Knee Injury Detection using MRI with Efficiently-Layered Network (ELNet)
May 06, 2020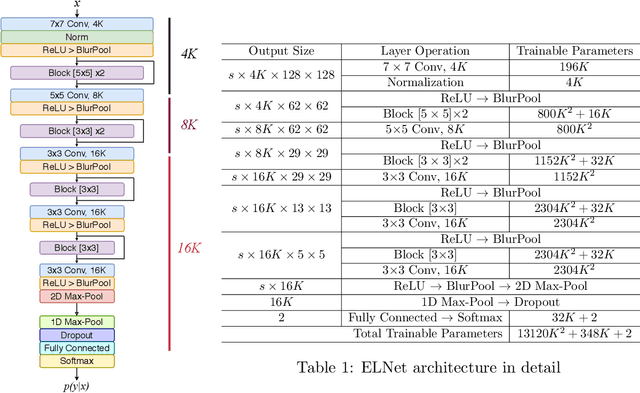
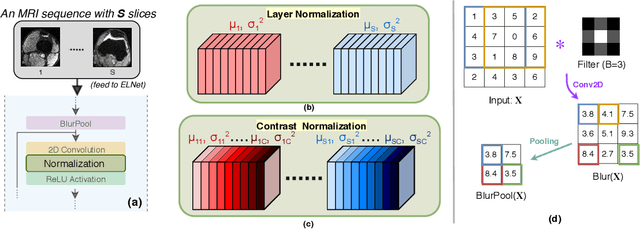
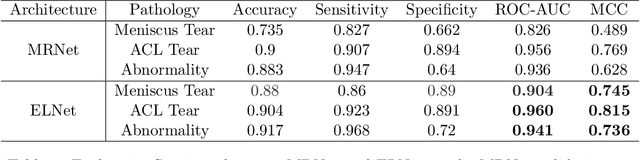
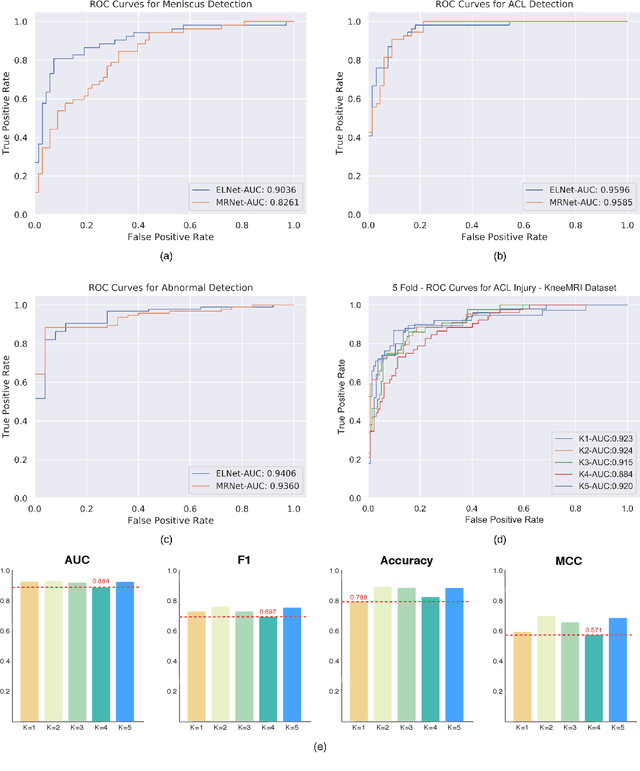
Abstract:Magnetic Resonance Imaging (MRI) is a widely-accepted imaging technique for knee injury analysis. Its advantage of capturing knee structure in three dimensions makes it the ideal tool for radiologists to locate potential tears in the knee. In order to better confront the ever growing workload of musculoskeletal (MSK) radiologists, automated tools for patients' triage are becoming a real need, reducing delays in the reading of pathological cases. In this work, we present the Efficiently-Layered Network (ELNet), a convolutional neural network (CNN) architecture optimized for the task of initial knee MRI diagnosis for triage. Unlike past approaches, we train ELNet from scratch instead of using a transfer-learning approach. The proposed method is validated quantitatively and qualitatively, and compares favorably against state-of-the-art MRNet while using a single imaging stack (axial or coronal) as input. Additionally, we demonstrate our model's capability to locate tears in the knee despite the absence of localization information during training. Lastly, the proposed model is extremely lightweight ($<$ 1MB) and therefore easy to train and deploy in real clinical settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge