Bradley Segal
Bridging the Generalisation Gap: Synthetic Data Generation for Multi-Site Clinical Model Validation
Apr 29, 2025



Abstract:Ensuring the generalisability of clinical machine learning (ML) models across diverse healthcare settings remains a significant challenge due to variability in patient demographics, disease prevalence, and institutional practices. Existing model evaluation approaches often rely on real-world datasets, which are limited in availability, embed confounding biases, and lack the flexibility needed for systematic experimentation. Furthermore, while generative models aim for statistical realism, they often lack transparency and explicit control over factors driving distributional shifts. In this work, we propose a novel structured synthetic data framework designed for the controlled benchmarking of model robustness, fairness, and generalisability. Unlike approaches focused solely on mimicking observed data, our framework provides explicit control over the data generating process, including site-specific prevalence variations, hierarchical subgroup effects, and structured feature interactions. This enables targeted investigation into how models respond to specific distributional shifts and potential biases. Through controlled experiments, we demonstrate the framework's ability to isolate the impact of site variations, support fairness-aware audits, and reveal generalisation failures, particularly highlighting how model complexity interacts with site-specific effects. This work contributes a reproducible, interpretable, and configurable tool designed to advance the reliable deployment of ML in clinical settings.
Synthesising clinically realistic Chest X-rays using Generative Adversarial Networks
Oct 07, 2020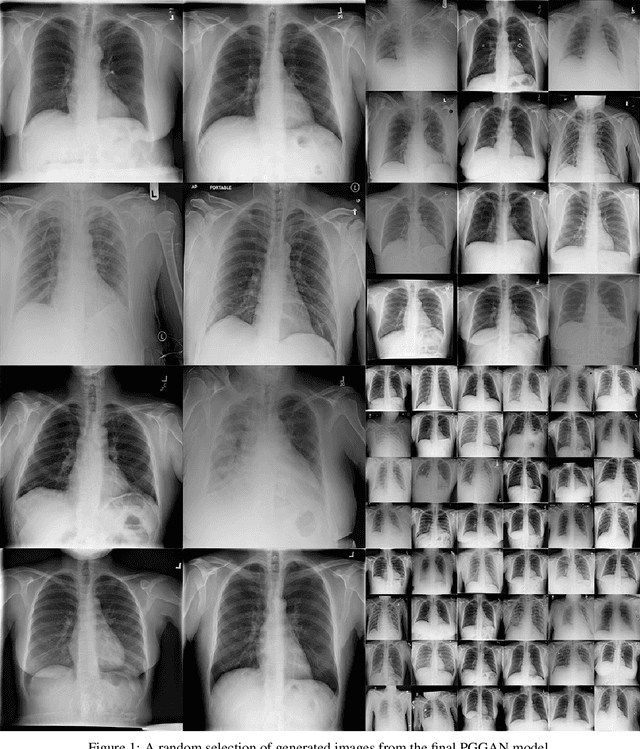
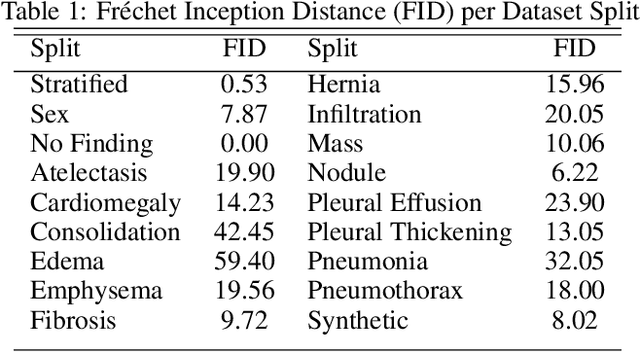
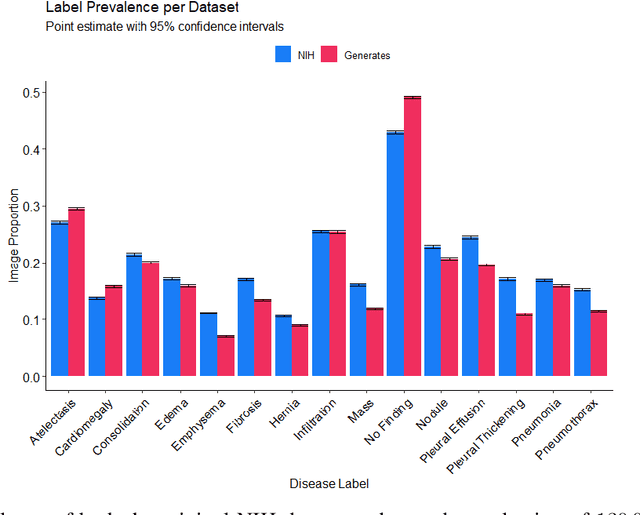
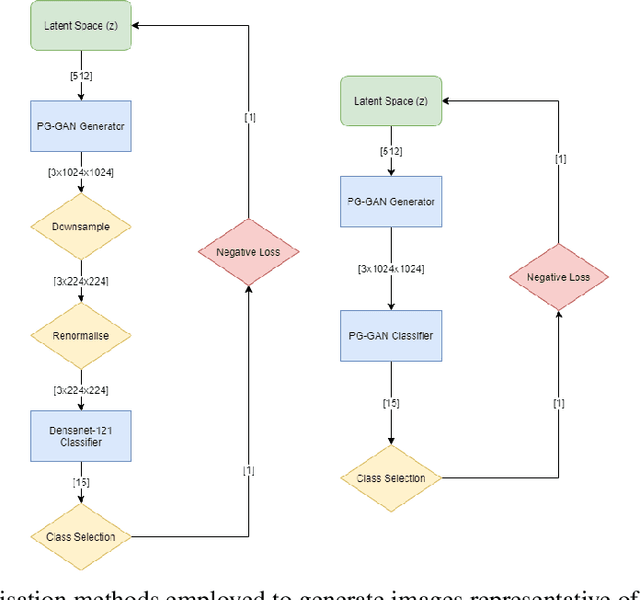
Abstract:Chest x-rays are one of the most commonly performed medical investigations globally and are vital to identifying a number of conditions. These images are however protected under patient confidentiality and as such require the removal of identifying information as well as ethical clearance to be released. Generative adversarial networks (GANs) are a branch of deep learning which are capable of producing synthetic samples of a desired distribution. Image generation is one such application with recent advances enabling the production of high-resolution images, a feature vital to the utility of x-rays given the scale of various pathologies. We apply the Progressive Growing GAN (PGGAN) to the task of chest x-ray generation with the goal of being able to produce images without any ethical concerns that may be used for medical education or in other machine learning work. We evaluate the properties of the generated x-rays with a practicing radiologist and demonstrate that high-quality, realistic images can be produced with global features consistent with pathologies seen in the NIH dataset. Improvements in the reproduction of small-scale details remains for future work. We train a classification model on the NIH images and evaluate the distribution of disease labels across the generated samples. We find that the model is capable of reproducing all the abnormalities in a similar proportion to the source image distribution as labelled by the classifier. We additionally demonstrate that the latent space can be optimised to produce images of a particular class despite unconditional training, with the model producing related features and complications for the class of interest. We also validate the application of the Fr'echet Inception Distance (FID) to x-ray images and determine that the PGGAN reproduces x-ray images with an FID of 8.02, which is similar to other high resolution tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge