Bradley J MacIntosh
Treatment-aware Diffusion Probabilistic Model for Longitudinal MRI Generation and Diffuse Glioma Growth Prediction
Sep 14, 2023



Abstract:Diffuse gliomas are malignant brain tumors that grow widespread through the brain. The complex interactions between neoplastic cells and normal tissue, as well as the treatment-induced changes often encountered, make glioma tumor growth modeling challenging. In this paper, we present a novel end-to-end network capable of generating future tumor masks and realistic MRIs of how the tumor will look at any future time points for different treatment plans. Our approach is based on cutting-edge diffusion probabilistic models and deep-segmentation neural networks. We included sequential multi-parametric magnetic resonance images (MRI) and treatment information as conditioning inputs to guide the generative diffusion process. This allows for tumor growth estimates at any given time point. We trained the model using real-world postoperative longitudinal MRI data with glioma tumor growth trajectories represented as tumor segmentation maps over time. The model has demonstrated promising performance across a range of tasks, including the generation of high-quality synthetic MRIs with tumor masks, time-series tumor segmentations, and uncertainty estimates. Combined with the treatment-aware generated MRIs, the tumor growth predictions with uncertainty estimates can provide useful information for clinical decision-making.
Voxels Intersecting along Orthogonal Levels Attention U-Net (viola-Unet) to Segment Intracerebral Haemorrhage Using Computed Tomography Head Scans
Aug 12, 2022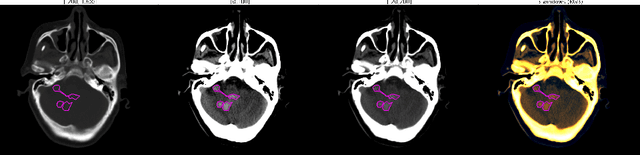
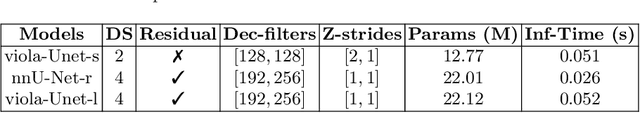
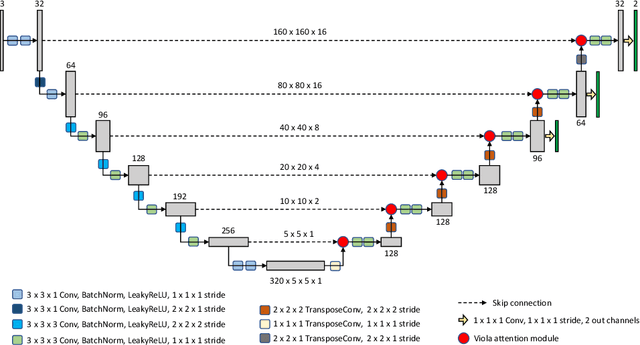
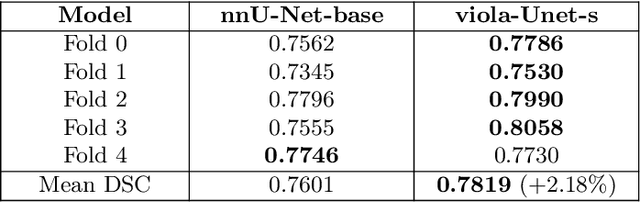
Abstract:We implemented two distinct 3-dimensional deep learning neural networks and evaluate their ability to segment intracranial hemorrhage (ICH) seen on non-contrast computed tomography (CT). One model, referred to as "Voxels-Intersecting along Orthogonal Levels of Attention U-Net" (viola-Unet), has architecture elements that are amenable to the INSTANCE 2022 Data Challenge. A second comparison model was derived from the no-new U-Net (nnU-Net). Input images and ground truth segmentation maps were used to train the two networks separately in supervised manner; validation data were subsequently used for semi-supervised training. Model predictions were compared during 5-fold cross validation. The viola-Unet outperformed the comparison network on two out of four performance metrics (i.e., NSD and RVD). An ensemble model that combined viola-Unet and nnU-Net networks had the highest performance for DSC and HD. We demonstrate there were ICH segmentation performance benefits associated with a 3D U-Net efficiently incorporates spatially orthogonal features during the decoding branch of the U-Net. The code base, pretrained weights, and docker image of the viola-Unet AI tool will be publicly available at https://github.com/samleoqh/Viola-Unet .
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge