Blaise Hanczar
IKrNet: A Neural Network for Detecting Specific Drug-Induced Patterns in Electrocardiograms Amidst Physiological Variability
May 12, 2025Abstract:Monitoring and analyzing electrocardiogram (ECG) signals, even under varying physiological conditions, including those influenced by physical activity, drugs and stress, is crucial to accurately assess cardiac health. However, current AI-based methods often fail to account for how these factors interact and alter ECG patterns, ultimately limiting their applicability in real-world settings. This study introduces IKrNet, a novel neural network model, which identifies drug-specific patterns in ECGs amidst certain physiological conditions. IKrNet's architecture incorporates spatial and temporal dynamics by using a convolutional backbone with varying receptive field size to capture spatial features. A bi-directional Long Short-Term Memory module is also employed to model temporal dependencies. By treating heart rate variability as a surrogate for physiological fluctuations, we evaluated IKrNet's performance across diverse scenarios, including conditions with physical stress, drug intake alone, and a baseline without drug presence. Our assessment follows a clinical protocol in which 990 healthy volunteers were administered 80mg of Sotalol, a drug which is known to be a precursor to Torsades-de-Pointes, a life-threatening arrhythmia. We show that IKrNet outperforms state-of-the-art models' accuracy and stability in varying physiological conditions, underscoring its clinical viability.
SurvCaus : Representation Balancing for Survival Causal Inference
Mar 29, 2022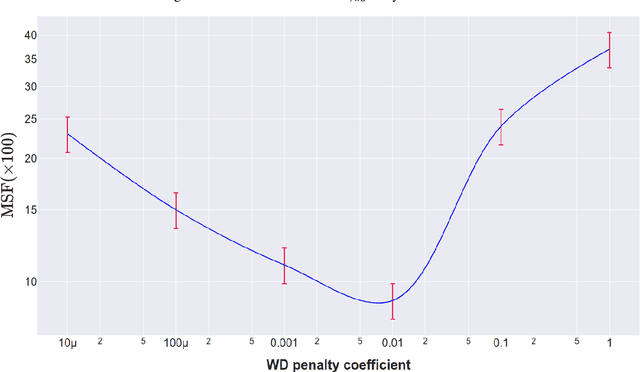
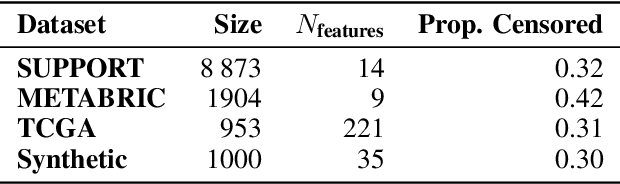
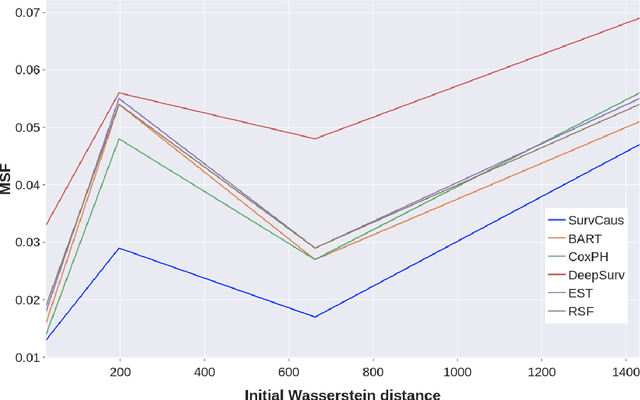

Abstract:Individual Treatment Effects (ITE) estimation methods have risen in popularity in the last years. Most of the time, individual effects are better presented as Conditional Average Treatment Effects (CATE). Recently, representation balancing techniques have gained considerable momentum in causal inference from observational data, still limited to continuous (and binary) outcomes. However, in numerous pathologies, the outcome of interest is a (possibly censored) survival time. Our paper proposes theoretical guarantees for a representation balancing framework applied to counterfactual inference in a survival setting using a neural network capable of predicting the factual and counterfactual survival functions (and then the CATE), in the presence of censorship, at the individual level. We also present extensive experiments on synthetic and semisynthetic datasets that show that the proposed extensions outperform baseline methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge