Bhagya Hettige
Temporal Cascade and Structural Modelling of EHRs for Granular Readmission Prediction
Feb 04, 2021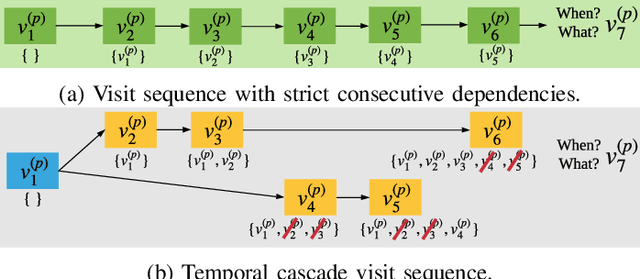
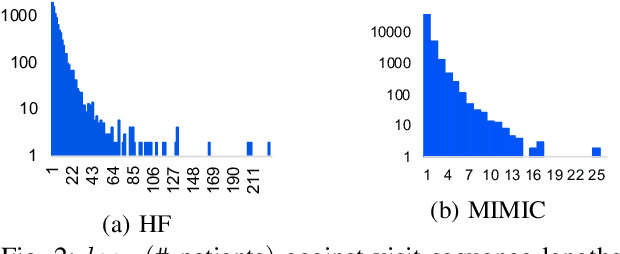
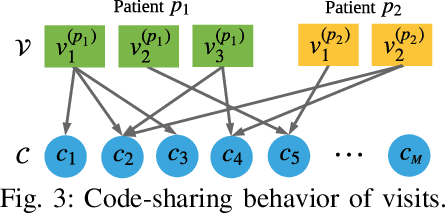
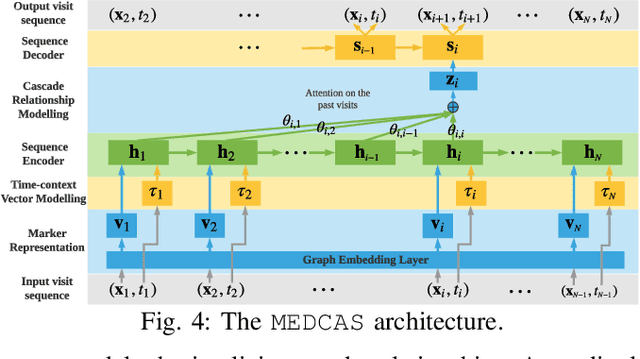
Abstract:Predicting (1) when the next hospital admission occurs and (2) what will happen in the next admission about a patient by mining electronic health record (EHR) data can provide granular readmission predictions to assist clinical decision making. Recurrent neural network (RNN) and point process models are usually employed in modelling temporal sequential data. Simple RNN models assume that sequences of hospital visits follow strict causal dependencies between consecutive visits. However, in the real-world, a patient may have multiple co-existing chronic medical conditions, i.e., multimorbidity, which results in a cascade of visits where a non-immediate historical visit can be most influential to the next visit. Although a point process (e.g., Hawkes process) is able to model a cascade temporal relationship, it strongly relies on a prior generative process assumption. We propose a novel model, MEDCAS, to address these challenges. MEDCAS combines the strengths of RNN-based models and point processes by integrating point processes in modelling visit types and time gaps into an attention-based sequence-to-sequence learning model, which is able to capture the temporal cascade relationships. To supplement the patients with short visit sequences, a structural modelling technique with graph-based methods is used to construct the markers of the point process in MEDCAS. Extensive experiments on three real-world EHR datasets have been performed and the results demonstrate that \texttt{MEDCAS} outperforms state-of-the-art models in both tasks.
$\mathtt{MedGraph:}$ Structural and Temporal Representation Learning of Electronic Medical Records
Dec 08, 2019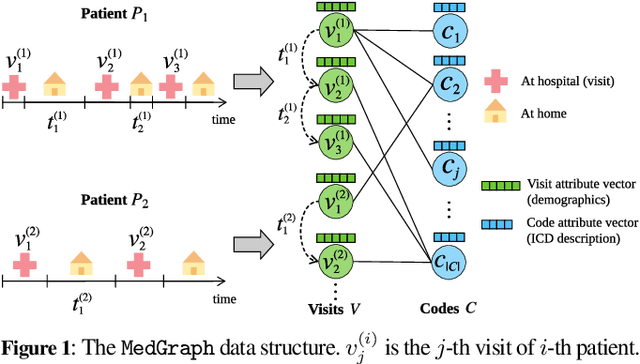
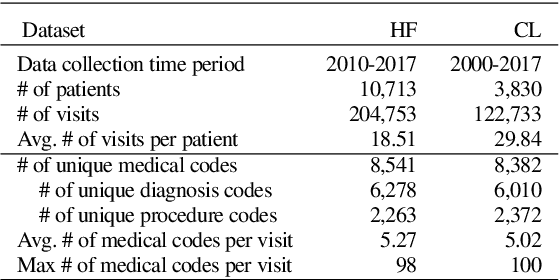
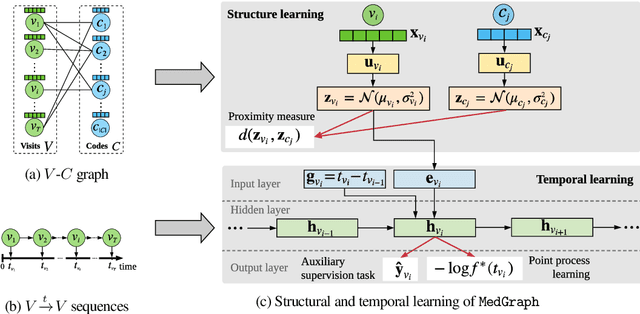
Abstract:Electronic medical record (EMR) data contains historical sequences of visits of patients, and each visit contains rich information, such as patient demographics, hospital utilisation and medical codes, including diagnosis, procedure and medication codes. Most existing EMR embedding methods capture visit-code associations by constructing input visit representations as binary vectors with a static vocabulary of medical codes. With this limited representation, they fail in encapsulating rich attribute information of visits (demographics and utilisation information) and/or codes (e.g., medical code descriptions). Furthermore, current work considers visits of the same patient as discrete-time events and ignores time gaps between them. However, the time gaps between visits depict dynamics of the patient's medical history inducing varying influences on future visits. To address these limitations, we present $\mathtt{MedGraph}$, a supervised EMR embedding method that captures two types of information: (1) the visit-code associations in an attributed bipartite graph, and (2) the temporal sequencing of visits through point processes. $\mathtt{MedGraph}$ produces Gaussian embeddings for visits and codes to model the uncertainty. We evaluate the performance of $\mathtt{MedGraph}$ through an extensive experimental study and show that $\mathtt{MedGraph}$ outperforms state-of-the-art EMR embedding methods in several medical risk prediction tasks.
Gaussian Embedding of Large-scale Attributed Graphs
Dec 02, 2019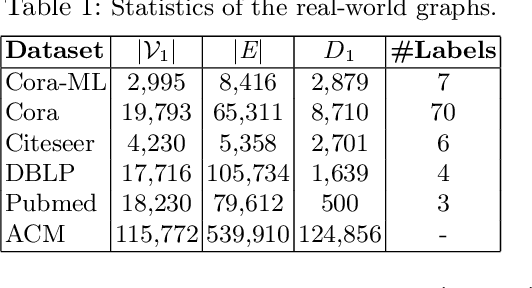
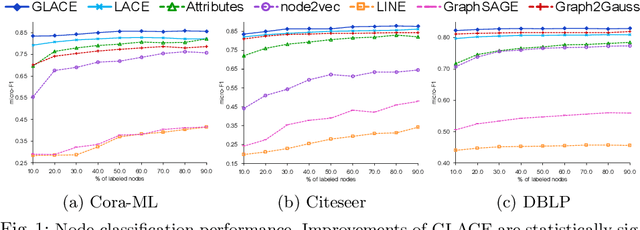
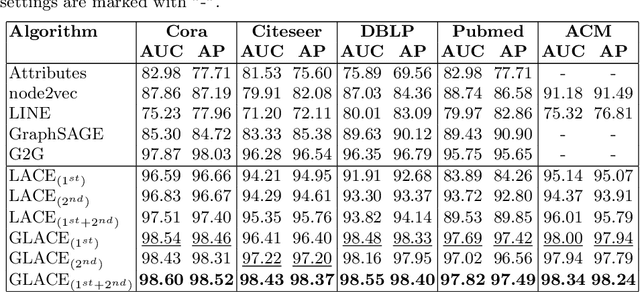
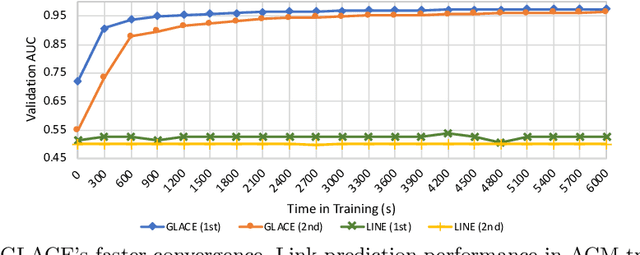
Abstract:Graph embedding methods transform high-dimensional and complex graph contents into low-dimensional representations. They are useful for a wide range of graph analysis tasks including link prediction, node classification, recommendation and visualization. Most existing approaches represent graph nodes as point vectors in a low-dimensional embedding space, ignoring the uncertainty present in the real-world graphs. Furthermore, many real-world graphs are large-scale and rich in content (e.g. node attributes). In this work, we propose GLACE, a novel, scalable graph embedding method that preserves both graph structure and node attributes effectively and efficiently in an end-to-end manner. GLACE effectively models uncertainty through Gaussian embeddings, and supports inductive inference of new nodes based on their attributes. In our comprehensive experiments, we evaluate GLACE on real-world graphs, and the results demonstrate that GLACE significantly outperforms state-of-the-art embedding methods on multiple graph analysis tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge