Benjamin B. Machta
In-Context Learning can distort the relationship between sequence likelihoods and biological fitness
Apr 23, 2025



Abstract:Language models have emerged as powerful predictors of the viability of biological sequences. During training these models learn the rules of the grammar obeyed by sequences of amino acids or nucleotides. Once trained, these models can take a sequence as input and produce a likelihood score as an output; a higher likelihood implies adherence to the learned grammar and correlates with experimental fitness measurements. Here we show that in-context learning can distort the relationship between fitness and likelihood scores of sequences. This phenomenon most prominently manifests as anomalously high likelihood scores for sequences that contain repeated motifs. We use protein language models with different architectures trained on the masked language modeling objective for our experiments, and find transformer-based models to be particularly vulnerable to this effect. This behavior is mediated by a look-up operation where the model seeks the identity of the masked position by using the other copy of the repeated motif as a reference. This retrieval behavior can override the model's learned priors. This phenomenon persists for imperfectly repeated sequences, and extends to other kinds of biologically relevant features such as reversed complement motifs in RNA sequences that fold into hairpin structures.
Far from Asymptopia
May 06, 2022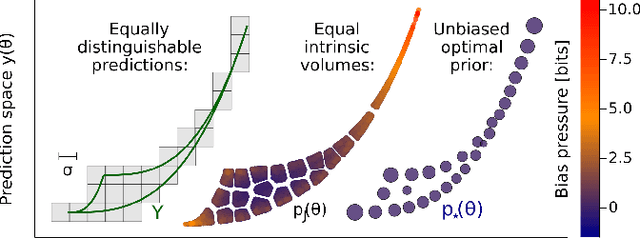
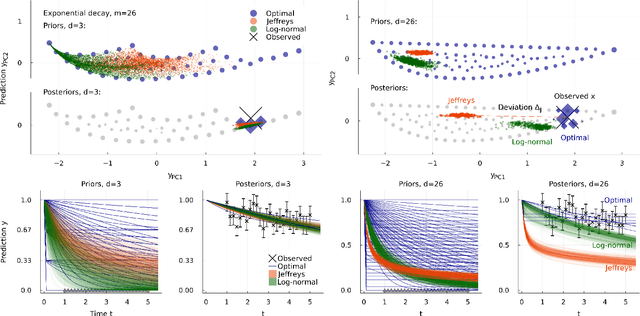
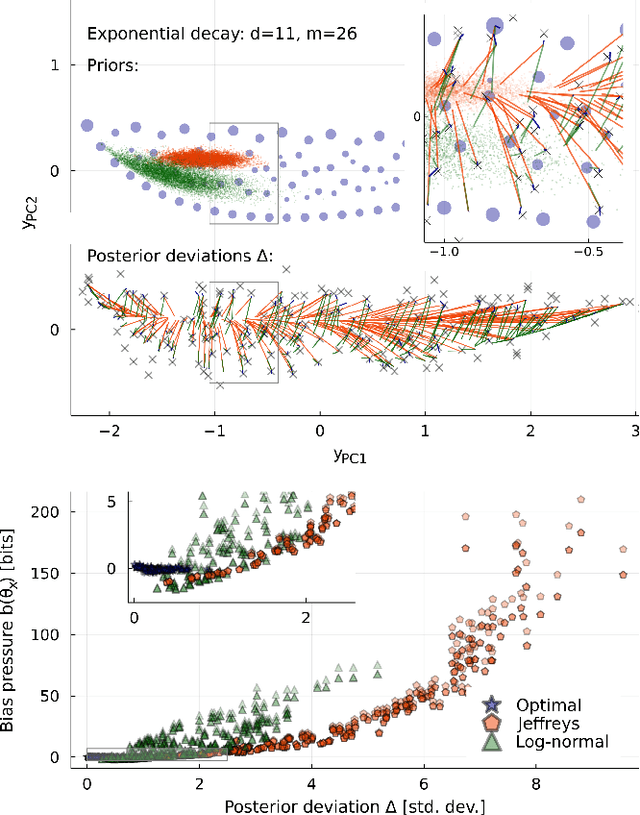
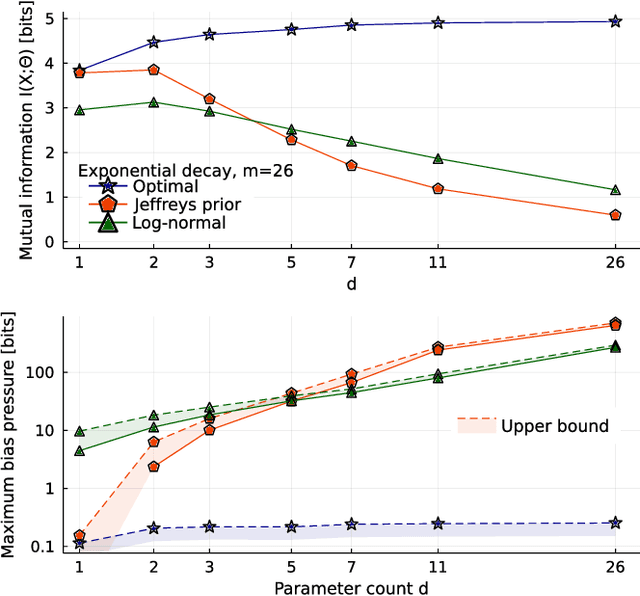
Abstract:Inference from limited data requires a notion of measure on parameter space, which is most explicit in the Bayesian framework as a prior distribution. Jeffreys prior is the best-known uninformative choice, the invariant volume element from information geometry, but we demonstrate here that this leads to enormous bias in typical high-dimensional models. This is because models found in science typically have an effective dimensionality of accessible behaviours much smaller than the number of microscopic parameters. Any measure which treats all of these parameters equally is far from uniform when projected onto the sub-space of relevant parameters, due to variations in the local co-volume of irrelevant directions. We present results on a principled choice of measure which avoids this issue, and leads to unbiased posteriors, by focusing on relevant parameters. This optimal prior depends on the quantity of data to be gathered, and approaches Jeffreys prior in the asymptotic limit. But for typical models this limit cannot be justified without an impossibly large increase in the quantity of data, exponential in the number of microscopic parameters.
Maximizing the information learned from finite data selects a simple model
Feb 14, 2018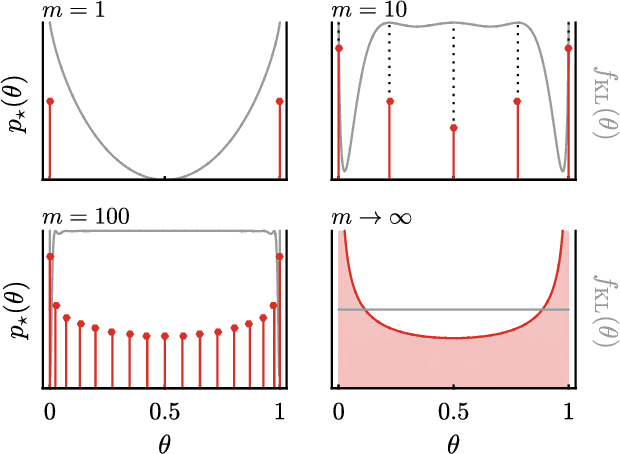
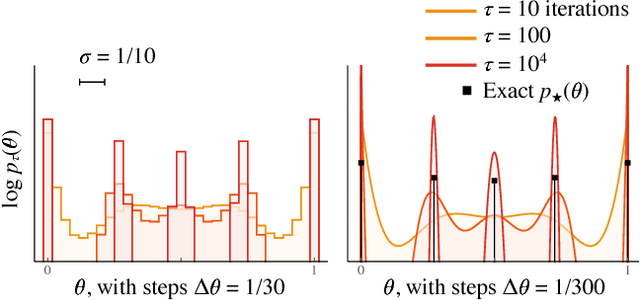
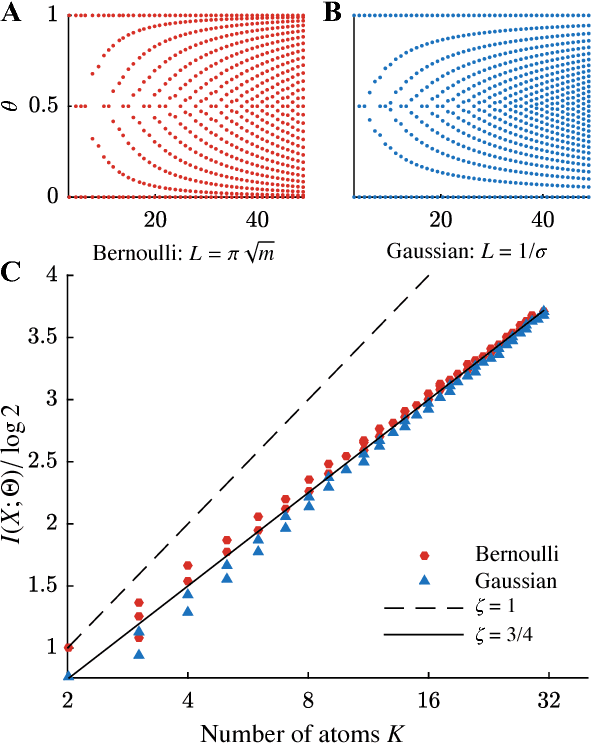
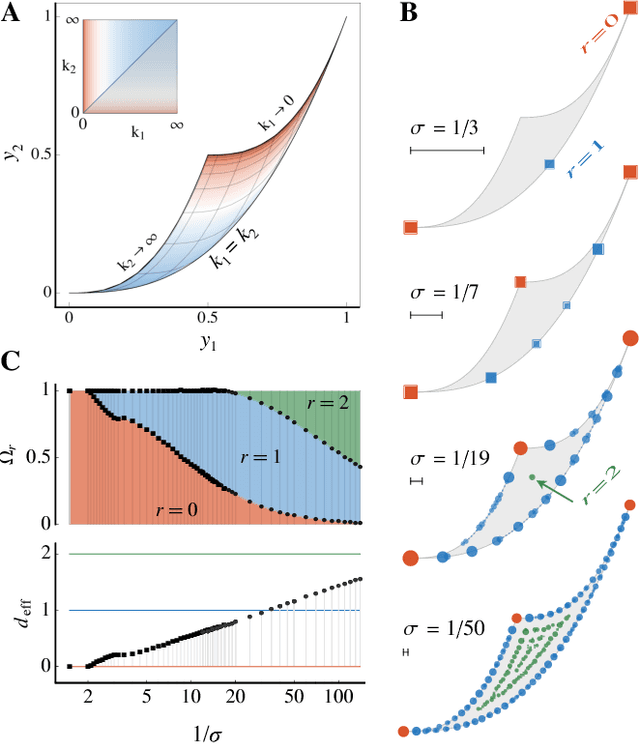
Abstract:We use the language of uninformative Bayesian prior choice to study the selection of appropriately simple effective models. We advocate for the prior which maximizes the mutual information between parameters and predictions, learning as much as possible from limited data. When many parameters are poorly constrained by the available data, we find that this prior puts weight only on boundaries of the parameter manifold. Thus it selects a lower-dimensional effective theory in a principled way, ignoring irrelevant parameter directions. In the limit where there is sufficient data to tightly constrain any number of parameters, this reduces to Jeffreys prior. But we argue that this limit is pathological when applied to the hyper-ribbon parameter manifolds generic in science, because it leads to dramatic dependence on effects invisible to experiment.
* 9 pages, 8 figures. v3 has improved discussion and adds an appendix about MDL and Bayes factors, and matches version to appear in PNAS (modulo comma placement). Title changed from "Rational Ignorance: Simpler Models Learn More Information from Finite Data"
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge