Anil Bharath
Architectural Exploration of Hybrid Neural Decoders for Neuromorphic Implantable BMI
May 09, 2025Abstract:This work presents an efficient decoding pipeline for neuromorphic implantable brain-machine interfaces (Neu-iBMI), leveraging sparse neural event data from an event-based neural sensing scheme. We introduce a tunable event filter (EvFilter), which also functions as a spike detector (EvFilter-SPD), significantly reducing the number of events processed for decoding by 192X and 554X, respectively. The proposed pipeline achieves high decoding performance, up to R^2=0.73, with ANN- and SNN-based decoders, eliminating the need for signal recovery, spike detection, or sorting, commonly performed in conventional iBMI systems. The SNN-Decoder reduces computations and memory required by 5-23X compared to NN-, and LSTM-Decoders, while the ST-NN-Decoder delivers similar performance to an LSTM-Decoder requiring 2.5X fewer resources. This streamlined approach significantly reduces computational and memory demands, making it ideal for low-power, on-implant, or wearable iBMIs.
Disentangling ODE parameters from dynamics in VAEs
Aug 26, 2021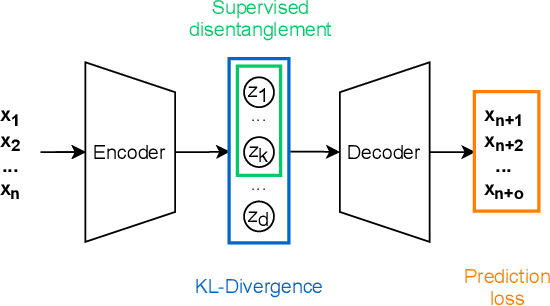
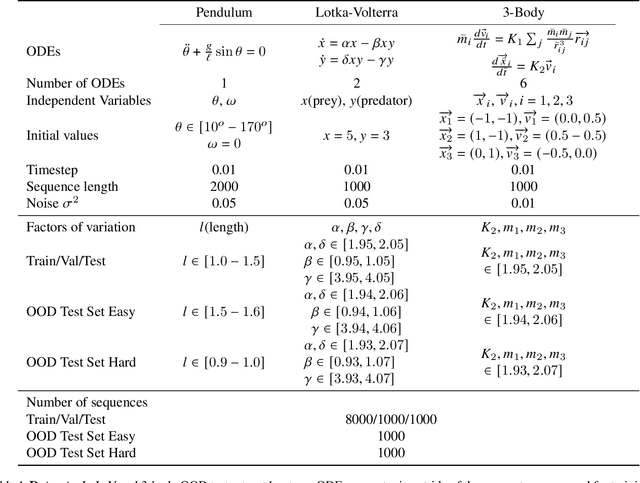
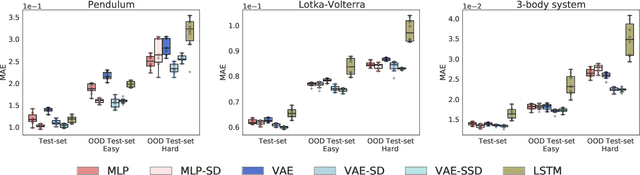
Abstract:Deep networks have become increasingly of interest in dynamical system prediction, but generalization remains elusive. In this work, we consider the physical parameters of ODEs as factors of variation of the data generating process. By leveraging ideas from supervised disentanglement in VAEs, we aim to separate the ODE parameters from the dynamics in the latent space. Experiments show that supervised disentanglement allows VAEs to capture the variability in the dynamics and extrapolate better to ODE parameter spaces that were not present in the training data.
Simulating Surface Wave Dynamics with Convolutional Networks
Dec 01, 2020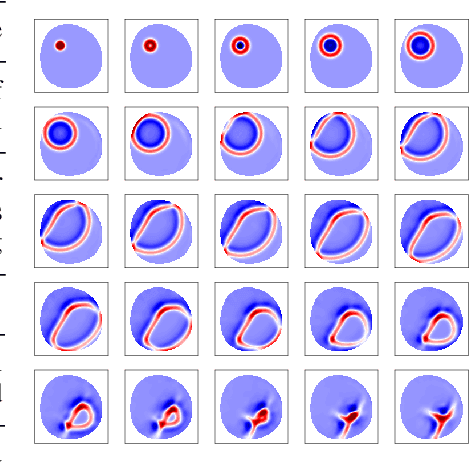
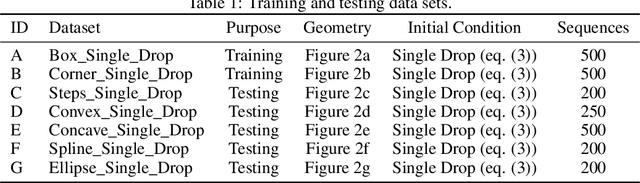
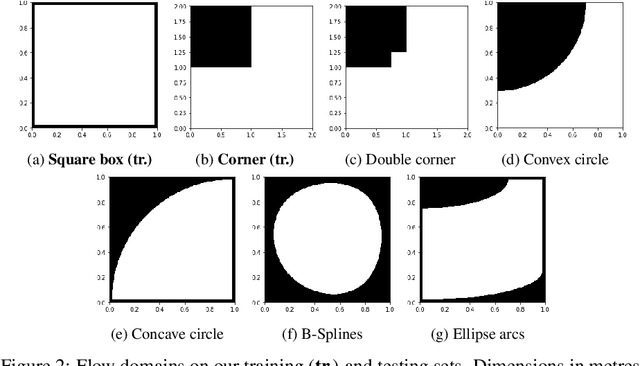
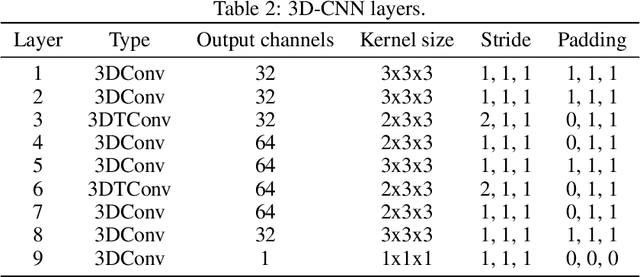
Abstract:We investigate the performance of fully convolutional networks to simulate the motion and interaction of surface waves in open and closed complex geometries. We focus on a U-Net architecture and analyse how well it generalises to geometric configurations not seen during training. We demonstrate that a modified U-Net architecture is capable of accurately predicting the height distribution of waves on a liquid surface within curved and multi-faceted open and closed geometries, when only simple box and right-angled corner geometries were seen during training. We also consider a separate and independent 3D CNN for performing time-interpolation on the predictions produced by our U-Net. This allows generating simulations with a smaller time-step size than the one the U-Net has been trained for.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge