Alzayat Saleh
Transformer-based Self-Supervised Fish Segmentation in Underwater Videos
Jun 11, 2022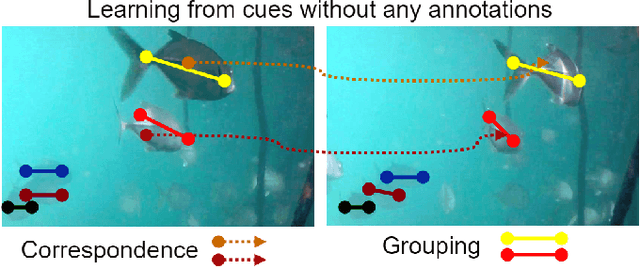
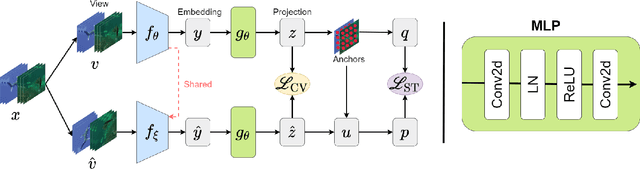
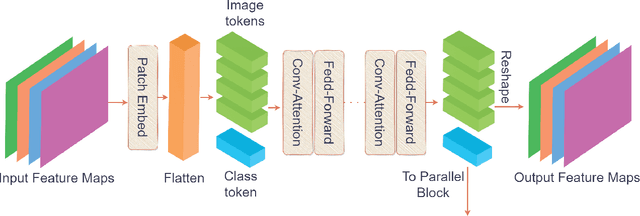
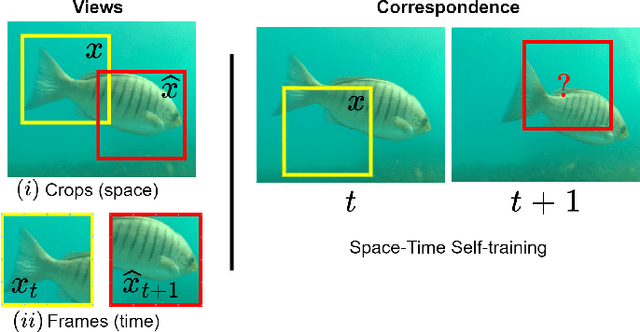
Abstract:Underwater fish segmentation to estimate fish body measurements is still largely unsolved due to the complex underwater environment. Relying on fully-supervised segmentation models requires collecting per-pixel labels, which is time-consuming and prone to overfitting. Self-supervised learning methods can help avoid the requirement of large annotated training datasets, however, to be useful in real-world applications, they should achieve good segmentation quality. In this paper, we introduce a Transformer-based method that uses self-supervision for high-quality fish segmentation. Our proposed model is trained on videos -- without any annotations -- to perform fish segmentation in underwater videos taken in situ in the wild. We show that when trained on a set of underwater videos from one dataset, the proposed model surpasses previous CNN-based and Transformer-based self-supervised methods and achieves performance relatively close to supervised methods on two new unseen underwater video datasets. This demonstrates the great generalisability of our model and the fact that it does not need a pre-trained model. In addition, we show that, due to its dense representation learning, our model is compute-efficient. We provide quantitative and qualitative results that demonstrate our model's significant capabilities.
Computer Vision and Deep Learning for Fish Classification in Underwater Habitats: A Survey
Mar 15, 2022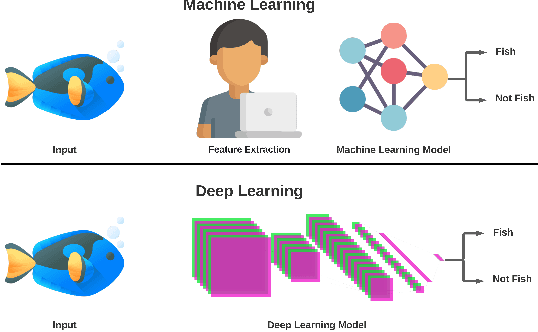
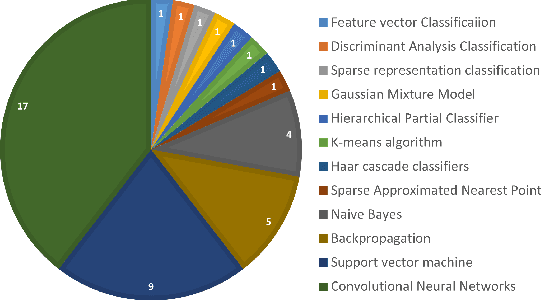
Abstract:Marine scientists use remote underwater video recording to survey fish species in their natural habitats. This helps them understand and predict how fish respond to climate change, habitat degradation, and fishing pressure. This information is essential for developing sustainable fisheries for human consumption, and for preserving the environment. However, the enormous volume of collected videos makes extracting useful information a daunting and time-consuming task for a human. A promising method to address this problem is the cutting-edge Deep Learning (DL) technology.DL can help marine scientists parse large volumes of video promptly and efficiently, unlocking niche information that cannot be obtained using conventional manual monitoring methods. In this paper, we provide an overview of the key concepts of DL, while presenting a survey of literature on fish habitat monitoring with a focus on underwater fish classification. We also discuss the main challenges faced when developing DL for underwater image processing and propose approaches to address them. Finally, we provide insights into the marine habitat monitoring research domain and shed light on what the future of DL for underwater image processing may hold. This paper aims to inform a wide range of readers from marine scientists who would like to apply DL in their research to computer scientists who would like to survey state-of-the-art DL-based underwater fish habitat monitoring literature.
A Deep Learning Localization Method for Measuring Abdominal Muscle Dimensions in Ultrasound Images
Sep 30, 2021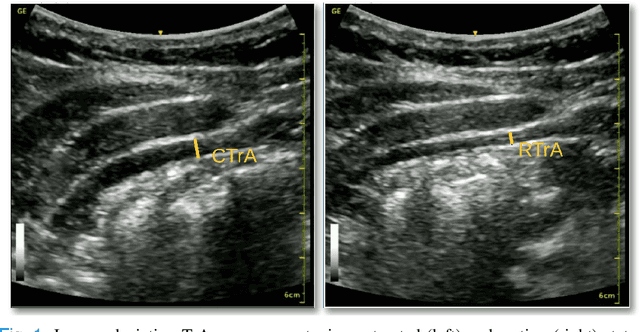
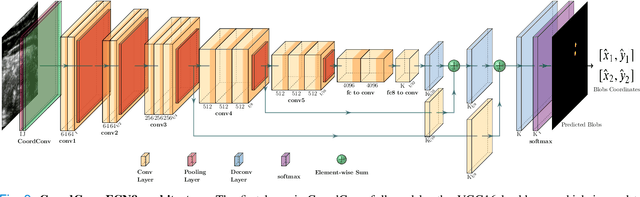

Abstract:Health professionals extensively use Two- Dimensional (2D) Ultrasound (US) videos and images to visualize and measure internal organs for various purposes including evaluation of muscle architectural changes. US images can be used to measure abdominal muscles dimensions for the diagnosis and creation of customized treatment plans for patients with Low Back Pain (LBP), however, they are difficult to interpret. Due to high variability, skilled professionals with specialized training are required to take measurements to avoid low intra-observer reliability. This variability stems from the challenging nature of accurately finding the correct spatial location of measurement endpoints in abdominal US images. In this paper, we use a Deep Learning (DL) approach to automate the measurement of the abdominal muscle thickness in 2D US images. By treating the problem as a localization task, we develop a modified Fully Convolutional Network (FCN) architecture to generate blobs of coordinate locations of measurement endpoints, similar to what a human operator does. We demonstrate that using the TrA400 US image dataset, our network achieves a Mean Absolute Error (MAE) of 0.3125 on the test set, which almost matches the performance of skilled ultrasound technicians. Our approach can facilitate next steps for automating the process of measurements in 2D US images, while reducing inter-observer as well as intra-observer variability for more effective clinical outcomes.
Affinity LCFCN: Learning to Segment Fish with Weak Supervision
Nov 06, 2020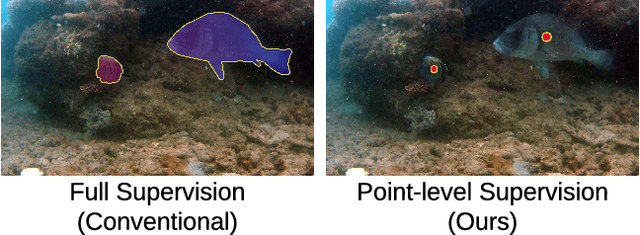
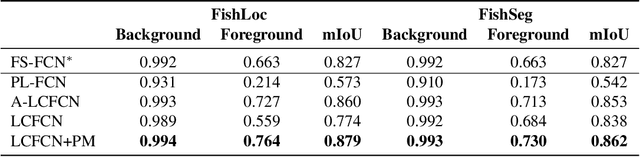

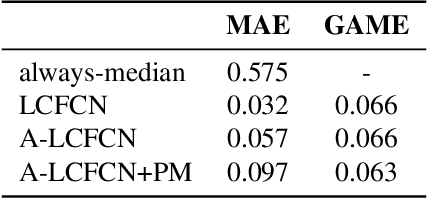
Abstract:Aquaculture industries rely on the availability of accurate fish body measurements, e.g., length, width and mass. Manual methods that rely on physical tools like rulers are time and labour intensive. Leading automatic approaches rely on fully-supervised segmentation models to acquire these measurements but these require collecting per-pixel labels -- also time consuming and laborious: i.e., it can take up to two minutes per fish to generate accurate segmentation labels, almost always requiring at least some manual intervention. We propose an automatic segmentation model efficiently trained on images labeled with only point-level supervision, where each fish is annotated with a single click. This labeling process requires significantly less manual intervention, averaging roughly one second per fish. Our approach uses a fully convolutional neural network with one branch that outputs per-pixel scores and another that outputs an affinity matrix. We aggregate these two outputs using a random walk to obtain the final, refined per-pixel segmentation output. We train the entire model end-to-end with an LCFCN loss, resulting in our A-LCFCN method. We validate our model on the DeepFish dataset, which contains many fish habitats from the north-eastern Australian region. Our experimental results confirm that A-LCFCN outperforms a fully-supervised segmentation model at fixed annotation budget. Moreover, we show that A-LCFCN achieves better segmentation results than LCFCN and a standard baseline. We have released the code at \url{https://github.com/IssamLaradji/affinity_lcfcn}.
A Realistic Fish-Habitat Dataset to Evaluate Algorithms for Underwater Visual Analysis
Aug 28, 2020
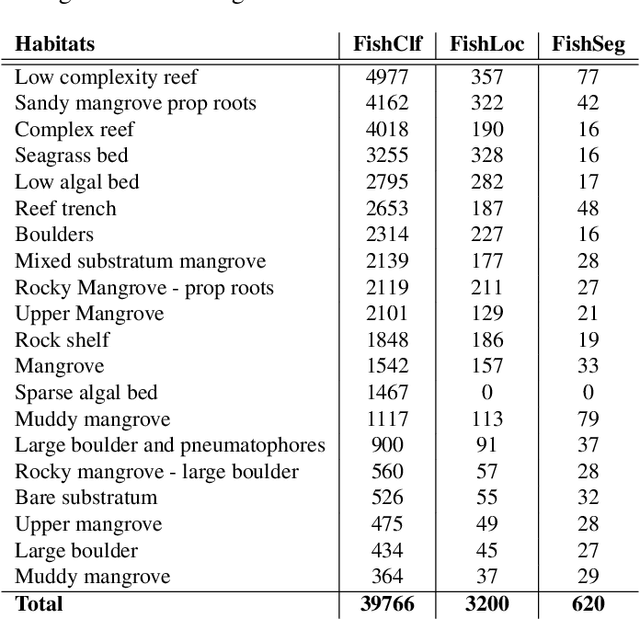
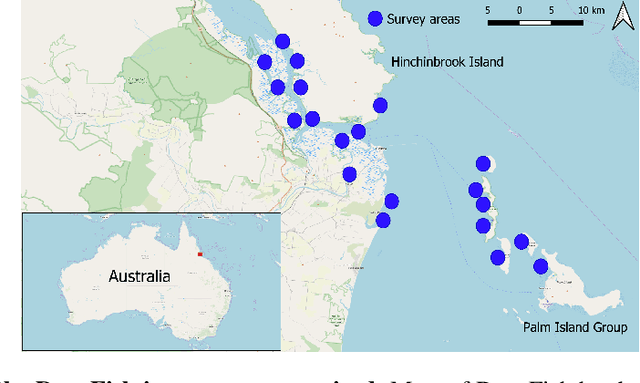

Abstract:Visual analysis of complex fish habitats is an important step towards sustainable fisheries for human consumption and environmental protection. Deep Learning methods have shown great promise for scene analysis when trained on large-scale datasets. However, current datasets for fish analysis tend to focus on the classification task within constrained, plain environments which do not capture the complexity of underwater fish habitats. To address this limitation, we present DeepFish as a benchmark suite with a large-scale dataset to train and test methods for several computer vision tasks. The dataset consists of approximately 40 thousand images collected underwater from 20 \green{habitats in the} marine-environments of tropical Australia. The dataset originally contained only classification labels. Thus, we collected point-level and segmentation labels to have a more comprehensive fish analysis benchmark. These labels enable models to learn to automatically monitor fish count, identify their locations, and estimate their sizes. Our experiments provide an in-depth analysis of the dataset characteristics, and the performance evaluation of several state-of-the-art approaches based on our benchmark. Although models pre-trained on ImageNet have successfully performed on this benchmark, there is still room for improvement. Therefore, this benchmark serves as a testbed to motivate further development in this challenging domain of underwater computer vision. Code is available at: https://github.com/alzayats/DeepFish
Automatic Weight Estimation of Harvested Fish from Images
Sep 06, 2019

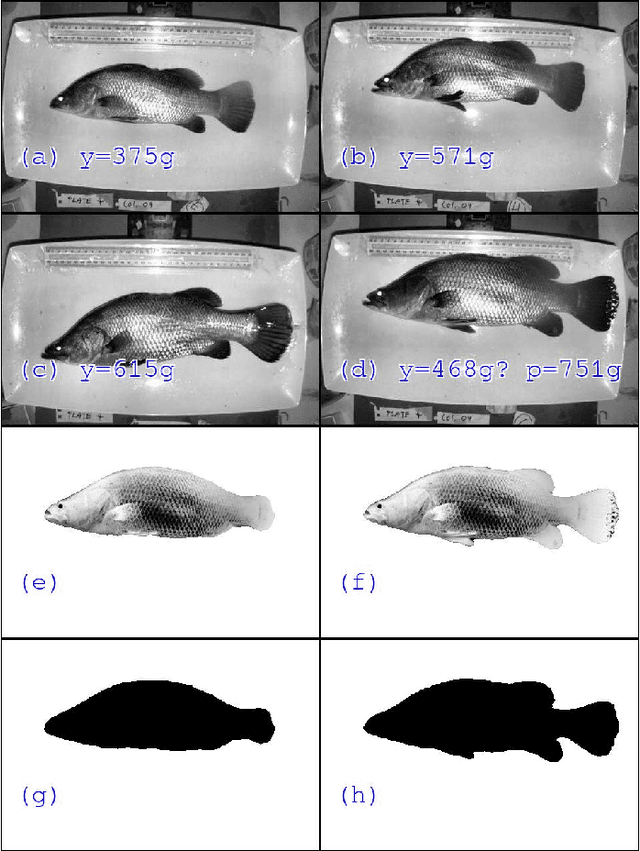
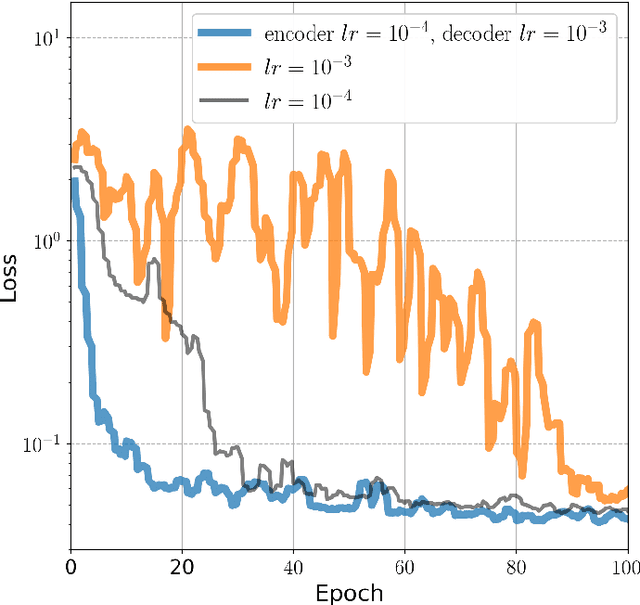
Abstract:Approximately 2,500 weights and corresponding images of harvested Lates calcarifer (Asian seabass or barramundi) were collected at three different locations in Queensland, Australia. Two instances of the LinkNet-34 segmentation Convolutional Neural Network (CNN) were trained. The first one was trained on 200 manually segmented fish masks with excluded fins and tails. The second was trained on 100 whole-fish masks. The two CNNs were applied to the rest of the images and yielded automatically segmented masks. The one-factor and two-factor simple mathematical weight-from-area models were fitted on 1072 area-weight pairs from the first two locations, where area values were extracted from the automatically segmented masks. When applied to 1,400 test images (from the third location), the one-factor whole-fish mask model achieved the best mean absolute percentage error (MAPE), MAPE=4.36%. Direct weight-from-image regression CNNs were also trained, where the no-fins based CNN performed best on the test images with MAPE=4.28%.
Underwater Fish Detection with Weak Multi-Domain Supervision
May 26, 2019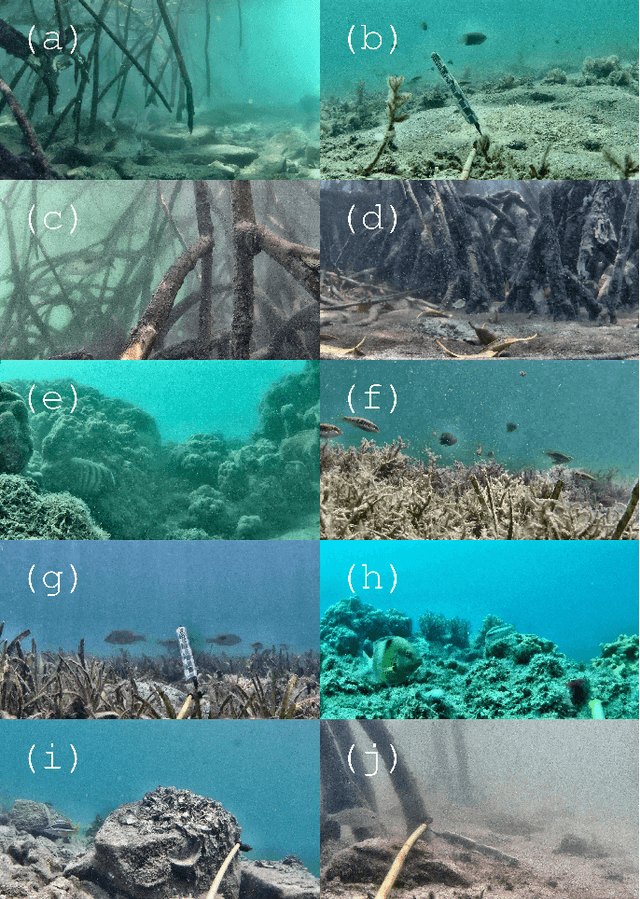
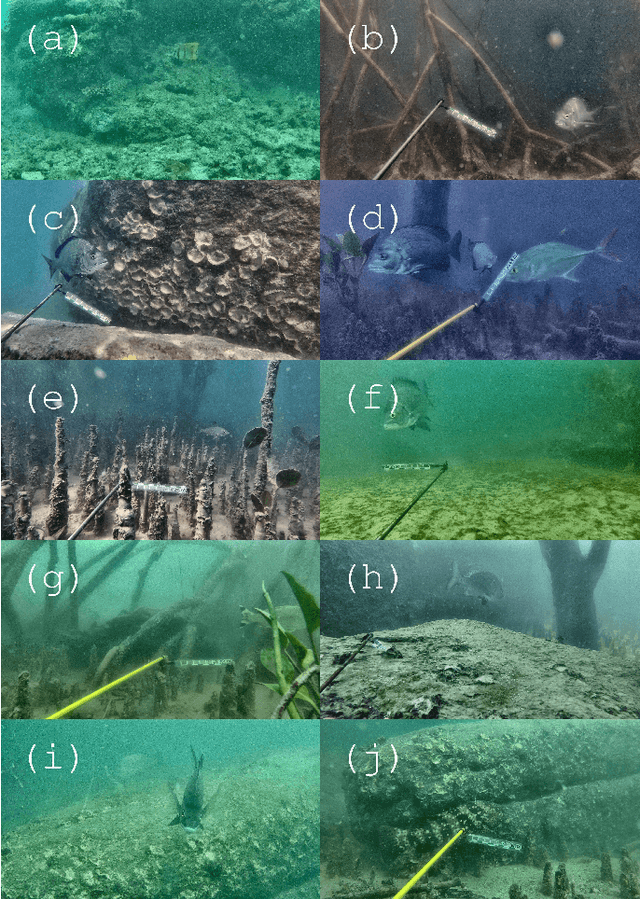


Abstract:Given a sufficiently large training dataset, it is relatively easy to train a modern convolution neural network (CNN) as a required image classifier. However, for the task of fish classification and/or fish detection, if a CNN was trained to detect or classify particular fish species in particular background habitats, the same CNN exhibits much lower accuracy when applied to new/unseen fish species and/or fish habitats. Therefore, in practice, the CNN needs to be continuously fine-tuned to improve its classification accuracy to handle new project-specific fish species or habitats. In this work we present a labelling-efficient method of training a CNN-based fish-detector (the Xception CNN was used as the base) on relatively small numbers (4,000) of project-domain underwater fish/no-fish images from 20 different habitats. Additionally, 17,000 of known negative (that is, missing fish) general-domain (VOC2012) above-water images were used. Two publicly available fish-domain datasets supplied additional 27,000 of above-water and underwater positive/fish images. By using this multi-domain collection of images, the trained Xception-based binary (fish/not-fish) classifier achieved 0.17% false-positives and 0.61% false-negatives on the project's 20,000 negative and 16,000 positive holdout test images, respectively. The area under the ROC curve (AUC) was 99.94%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge