Lung Nodule Classification
Papers and Code
Tri-Reader: An Open-Access, Multi-Stage AI Pipeline for First-Pass Lung Nodule Annotation in Screening CT
Jan 27, 2026Using multiple open-access models trained on public datasets, we developed Tri-Reader, a comprehensive, freely available pipeline that integrates lung segmentation, nodule detection, and malignancy classification into a unified tri-stage workflow. The pipeline is designed to prioritize sensitivity while reducing the candidate burden for annotators. To ensure accuracy and generalizability across diverse practices, we evaluated Tri-Reader on multiple internal and external datasets as compared with expert annotations and dataset-provided reference standards.
MedSAM-based lung masking for multi-label chest X-ray classification
Dec 28, 2025Chest X-ray (CXR) imaging is widely used for screening and diagnosing pulmonary abnormalities, yet automated interpretation remains challenging due to weak disease signals, dataset bias, and limited spatial supervision. Foundation models for medical image segmentation (MedSAM) provide an opportunity to introduce anatomically grounded priors that may improve robustness and interpretability in CXR analysis. We propose a segmentation-guided CXR classification pipeline that integrates MedSAM as a lung region extraction module prior to multi-label abnormality classification. MedSAM is fine-tuned using a public image-mask dataset from Airlangga University Hospital. We then apply it to a curated subset of the public NIH CXR dataset to train and evaluate deep convolutional neural networks for multi-label prediction of five abnormalities (Mass, Nodule, Pneumonia, Edema, and Fibrosis), with the normal case (No Finding) evaluated via a derived score. Experiments show that MedSAM produces anatomically plausible lung masks across diverse imaging conditions. We find that masking effects are both task-dependent and architecture-dependent. ResNet50 trained on original images achieves the strongest overall abnormality discrimination, while loose lung masking yields comparable macro AUROC but significantly improves No Finding discrimination, indicating a trade-off between abnormality-specific classification and normal case screening. Tight masking consistently reduces abnormality level performance but improves training efficiency. Loose masking partially mitigates this degradation by preserving perihilar and peripheral context. These results suggest that lung masking should be treated as a controllable spatial prior selected to match the backbone and clinical objective, rather than applied uniformly.
NodMAISI: Nodule-Oriented Medical AI for Synthetic Imaging
Dec 19, 2025Objective: Although medical imaging datasets are increasingly available, abnormal and annotation-intensive findings critical to lung cancer screening, particularly small pulmonary nodules, remain underrepresented and inconsistently curated. Methods: We introduce NodMAISI, an anatomically constrained, nodule-oriented CT synthesis and augmentation framework trained on a unified multi-source cohort (7,042 patients, 8,841 CTs, 14,444 nodules). The framework integrates: (i) a standardized curation and annotation pipeline linking each CT with organ masks and nodule-level annotations, (ii) a ControlNet-conditioned rectified-flow generator built on MAISI-v2's foundational blocks to enforce anatomy- and lesion-consistent synthesis, and (iii) lesion-aware augmentation that perturbs nodule masks (controlled shrinkage) while preserving surrounding anatomy to generate paired CT variants. Results: Across six public test datasets, NodMAISI improved distributional fidelity relative to MAISI-v2 (real-to-synthetic FID range 1.18 to 2.99 vs 1.69 to 5.21). In lesion detectability analysis using a MONAI nodule detector, NodMAISI substantially increased average sensitivity and more closely matched clinical scans (IMD-CT: 0.69 vs 0.39; DLCS24: 0.63 vs 0.20), with the largest gains for sub-centimeter nodules where MAISI-v2 frequently failed to reproduce the conditioned lesion. In downstream nodule-level malignancy classification trained on LUNA25 and externally evaluated on LUNA16, LNDbv4, and DLCS24, NodMAISI augmentation improved AUC by 0.07 to 0.11 at <=20% clinical data and by 0.12 to 0.21 at 10%, consistently narrowing the performance gap under data scarcity.
Minimum Data, Maximum Impact: 20 annotated samples for explainable lung nodule classification
Aug 01, 2025



Classification models that provide human-interpretable explanations enhance clinicians' trust and usability in medical image diagnosis. One research focus is the integration and prediction of pathology-related visual attributes used by radiologists alongside the diagnosis, aligning AI decision-making with clinical reasoning. Radiologists use attributes like shape and texture as established diagnostic criteria and mirroring these in AI decision-making both enhances transparency and enables explicit validation of model outputs. However, the adoption of such models is limited by the scarcity of large-scale medical image datasets annotated with these attributes. To address this challenge, we propose synthesizing attribute-annotated data using a generative model. We enhance the Diffusion Model with attribute conditioning and train it using only 20 attribute-labeled lung nodule samples from the LIDC-IDRI dataset. Incorporating its generated images into the training of an explainable model boosts performance, increasing attribute prediction accuracy by 13.4% and target prediction accuracy by 1.8% compared to training with only the small real attribute-annotated dataset. This work highlights the potential of synthetic data to overcome dataset limitations, enhancing the applicability of explainable models in medical image analysis.
Multi-Attention Stacked Ensemble for Lung Cancer Detection in CT Scans
Jul 27, 2025



In this work, we address the challenge of binary lung nodule classification (benign vs malignant) using CT images by proposing a multi-level attention stacked ensemble of deep neural networks. Three pretrained backbones - EfficientNet V2 S, MobileViT XXS, and DenseNet201 - are each adapted with a custom classification head tailored to 96 x 96 pixel inputs. A two-stage attention mechanism learns both model-wise and class-wise importance scores from concatenated logits, and a lightweight meta-learner refines the final prediction. To mitigate class imbalance and improve generalization, we employ dynamic focal loss with empirically calculated class weights, MixUp augmentation during training, and test-time augmentation at inference. Experiments on the LIDC-IDRI dataset demonstrate exceptional performance, achieving 98.09 accuracy and 0.9961 AUC, representing a 35 percent reduction in error rate compared to state-of-the-art methods. The model exhibits balanced performance across sensitivity (98.73) and specificity (98.96), with particularly strong results on challenging cases where radiologist disagreement was high. Statistical significance testing confirms the robustness of these improvements across multiple experimental runs. Our approach can serve as a robust, automated aid for radiologists in lung cancer screening.
Lung Nodule-SSM: Self-Supervised Lung Nodule Detection and Classification in Thoracic CT Images
May 21, 2025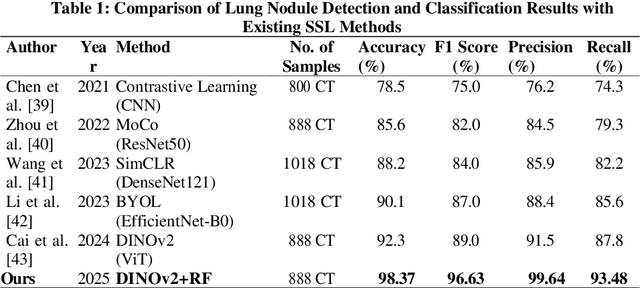
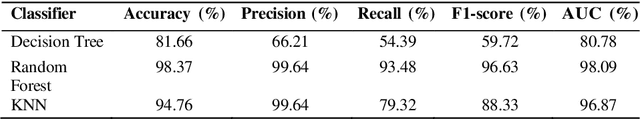
Lung cancer remains among the deadliest types of cancer in recent decades, and early lung nodule detection is crucial for improving patient outcomes. The limited availability of annotated medical imaging data remains a bottleneck in developing accurate computer-aided diagnosis (CAD) systems. Self-supervised learning can help leverage large amounts of unlabeled data to develop more robust CAD systems. With the recent advent of transformer-based architecture and their ability to generalize to unseen tasks, there has been an effort within the healthcare community to adapt them to various medical downstream tasks. Thus, we propose a novel "LungNodule-SSM" method, which utilizes selfsupervised learning with DINOv2 as a backbone to enhance lung nodule detection and classification without annotated data. Our methodology has two stages: firstly, the DINOv2 model is pre-trained on unlabeled CT scans to learn robust feature representations, then secondly, these features are fine-tuned using transformer-based architectures for lesionlevel detection and accurate lung nodule diagnosis. The proposed method has been evaluated on the challenging LUNA 16 dataset, consisting of 888 CT scans, and compared with SOTA methods. Our experimental results show the superiority of our proposed method with an accuracy of 98.37%, explaining its effectiveness in lung nodule detection. The source code, datasets, and pre-processed data can be accessed using the link:https://github.com/EMeRALDsNRPU/Lung-Nodule-SSM-Self-Supervised-Lung-Nodule-Detection-and-Classification/tree/main
A Residual Multi-task Network for Joint Classification and Regression in Medical Imaging
Feb 27, 2025



Detection and classification of pulmonary nodules is a challenge in medical image analysis due to the variety of shapes and sizes of nodules and their high concealment. Despite the success of traditional deep learning methods in image classification, deep networks still struggle to perfectly capture subtle changes in lung nodule detection. Therefore, we propose a residual multi-task network (Res-MTNet) model, which combines multi-task learning and residual learning, and improves feature representation ability by sharing feature extraction layer and introducing residual connections. Multi-task learning enables the model to handle multiple tasks simultaneously, while the residual module solves the problem of disappearing gradients, ensuring stable training of deeper networks and facilitating information sharing between tasks. Res-MTNet enhances the robustness and accuracy of the model, providing a more reliable lung nodule analysis tool for clinical medicine and telemedicine.
LMLCC-Net: A Semi-Supervised Deep Learning Model for Lung Nodule Malignancy Prediction from CT Scans using a Novel Hounsfield Unit-Based Intensity Filtering
May 09, 2025Lung cancer is the leading cause of patient mortality in the world. Early diagnosis of malignant pulmonary nodules in CT images can have a significant impact on reducing disease mortality and morbidity. In this work, we propose LMLCC-Net, a novel deep learning framework for classifying nodules from CT scan images using a 3D CNN, considering Hounsfield Unit (HU)-based intensity filtering. Benign and malignant nodules have significant differences in their intensity profile of HU, which was not exploited in the literature. Our method considers the intensity pattern as well as the texture for the prediction of malignancies. LMLCC-Net extracts features from multiple branches that each use a separate learnable HU-based intensity filtering stage. Various combinations of branches and learnable ranges of filters were explored to finally produce the best-performing model. In addition, we propose a semi-supervised learning scheme for labeling ambiguous cases and also developed a lightweight model to classify the nodules. The experimental evaluations are carried out on the LUNA16 dataset. Our proposed method achieves a classification accuracy (ACC) of 91.96%, a sensitivity (SEN) of 92.04%, and an area under the curve (AUC) of 91.87%, showing improved performance compared to existing methods. The proposed method can have a significant impact in helping radiologists in the classification of pulmonary nodules and improving patient care.
Advanced Lung Nodule Segmentation and Classification for Early Detection of Lung Cancer using SAM and Transfer Learning
Dec 31, 2024



Lung cancer is an extremely lethal disease primarily due to its late-stage diagnosis and significant mortality rate, making it the major cause of cancer-related demises globally. Machine Learning (ML) and Convolution Neural network (CNN) based Deep Learning (DL) techniques are primarily used for precise segmentation and classification of cancerous nodules in the CT (Computed Tomography) or MRI images. This study introduces an innovative approach to lung nodule segmentation by utilizing the Segment Anything Model (SAM) combined with transfer learning techniques. Precise segmentation of lung nodules is crucial for the early detection of lung cancer. The proposed method leverages Bounding Box prompts and a vision transformer model to enhance segmentation performance, achieving high accuracy, Dice Similarity Coefficient (DSC) and Intersection over Union (IoU) metrics. The integration of SAM and Transfer Learning significantly improves Computer-Aided Detection (CAD) systems in medical imaging, particularly for lung cancer diagnosis. The findings demonstrate the proposed model effectiveness in precisely segmenting lung nodules from CT scans, underscoring its potential to advance early detection and improve patient care outcomes in lung cancer diagnosis. The results show SAM Model with transfer learning achieving a DSC of 97.08% and an IoU of 95.6%, for segmentation and accuracy of 96.71% for classification indicates that ,its performance is noteworthy compared to existing techniques.
Hierarchical Vision Transformer with Prototypes for Interpretable Medical Image Classification
Feb 13, 2025



Explainability is a highly demanded requirement for applications in high-risk areas such as medicine. Vision Transformers have mainly been limited to attention extraction to provide insight into the model's reasoning. Our approach combines the high performance of Vision Transformers with the introduction of new explainability capabilities. We present HierViT, a Vision Transformer that is inherently interpretable and adapts its reasoning to that of humans. A hierarchical structure is used to process domain-specific features for prediction. It is interpretable by design, as it derives the target output with human-defined features that are visualized by exemplary images (prototypes). By incorporating domain knowledge about these decisive features, the reasoning is semantically similar to human reasoning and therefore intuitive. Moreover, attention heatmaps visualize the crucial regions for identifying each feature, thereby providing HierViT with a versatile tool for validating predictions. Evaluated on two medical benchmark datasets, LIDC-IDRI for lung nodule assessment and derm7pt for skin lesion classification, HierViT achieves superior and comparable prediction accuracy, respectively, while offering explanations that align with human reasoning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge