EEG-Based Emotion Recognition Using Regularized Graph Neural Networks
Paper and Code
Aug 26, 2019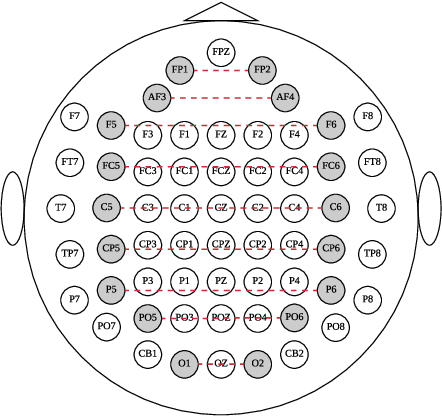
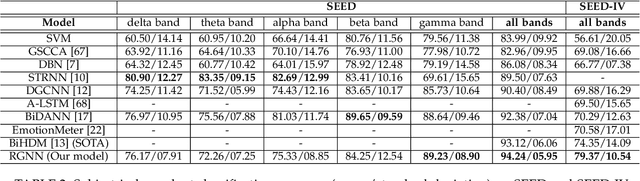
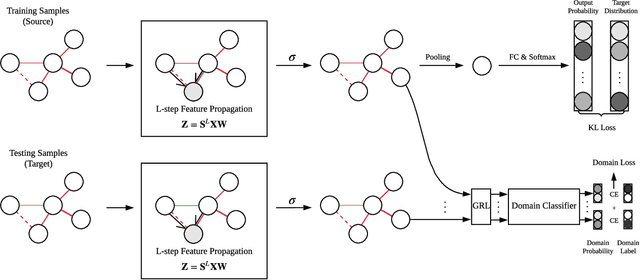
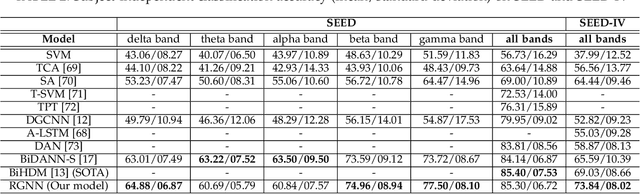
EEG signals measure the neuronal activities on different brain regions via electrodes. Many existing studies on EEG-based emotion recognition do not exploit the topological structure of EEG signals. In this paper, we propose a regularized graph neural network (RGNN) for EEG-based emotion recognition, which is biologically supported and captures both local and global inter-channel relations. Specifically, we model the inter-channel relations in EEG signals via an adjacency matrix in our graph neural network where the connection and sparseness of the adjacency matrix are supported by the neurosicience theories of human brain organization. In addition, we propose two regularizers, namely node-wise domain adversarial training (NodeDAT) and emotion-aware distribution learning (EmotionDL), to improve the robustness of our model against cross-subject EEG variations and noisy labels, respectively. To thoroughly evaluate our model, we conduct extensive experiments in both subject-dependent and subject-independent classification settings on two public datasets: SEED and SEED-IV. Our model obtains better performance than competitive baselines such as SVM, DBN, DGCNN, BiDANN, and the state-of-the-art BiHDM in most experimental settings . Our model analysis demonstrates that the proposed biologically supported adjacency matrix and two regularizers contribute consistent and significant gain to the performance. Investigations on the neuronal activities reveal that pre-frontal, parietal and occipital regions may be the most informative regions for emotion recognition, which is consistent with relevant prior studies. In addition, experimental results suggest that global inter-channel relations between the left and right hemispheres are important for emotion recognition and local inter-channel relations between (FP1, AF3), (F6, F8) and (FP2, AF4) may also provide useful information.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge