Xudong Fan
Machine-Learning Enabled Multidimensional Data Utilization in Multi-resonance Biosensors: A Pathway to Enhanced Accuracy
Dec 28, 2024Abstract:A novel framework is proposed that combines multi-resonance biosensors with machine learning (ML) to significantly enhance the accuracy of parameter prediction in biosensing. Unlike traditional single-resonance systems, which are limited to one-dimensional datasets, this approach leverages multi-dimensional data generated by a custom-designed nanostructure, a periodic array of silicon nanorods with a triangular cross-section over an aluminum reflector. High bulk sensitivity values are achieved for this multi-resonant structure, with certain resonant peaks reaching up to 1706 nm/RIU. The predictive power of multiple resonant peaks from transverse magnetic (TM) and transverse electric (TE) polarizations is evaluated using Ridge Regression modeling. Systematic analysis reveals that incorporating multiple resonances yields up to three orders of magnitude improvement in refractive index detection precision compared to single-peak analyses. This precision enhancement is achieved without modifications to the biosensor hardware, highlighting the potential of data-centric strategies in biosensing. The findings establish a new paradigm in biosensing, demonstrating that the synergy between multi-resonance data acquisition and ML-based analysis can significantly enhance detection accuracy. This study provides a scalable pathway for advancing high-precision biosensing technologies.
Retention time trajectory matching for target compound peak identification in chromatographic analysis
Jul 16, 2021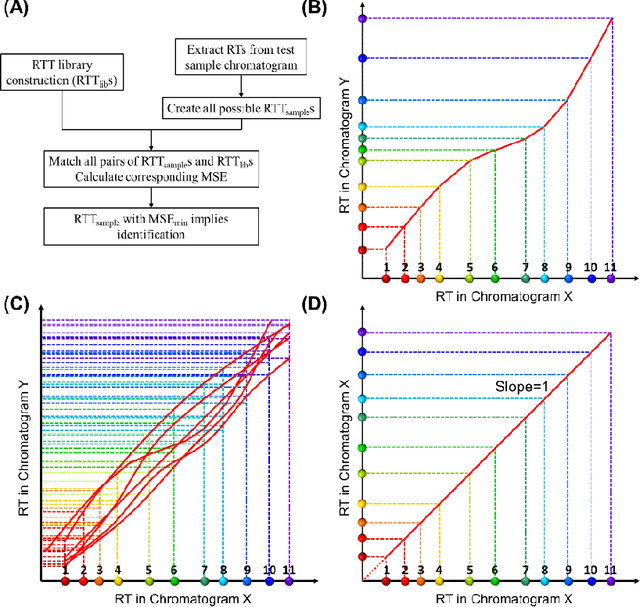
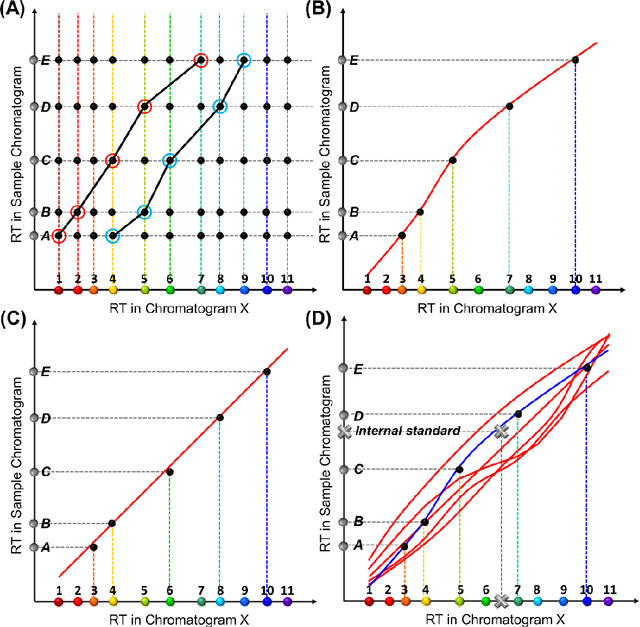
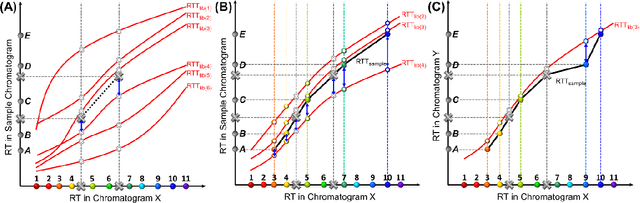
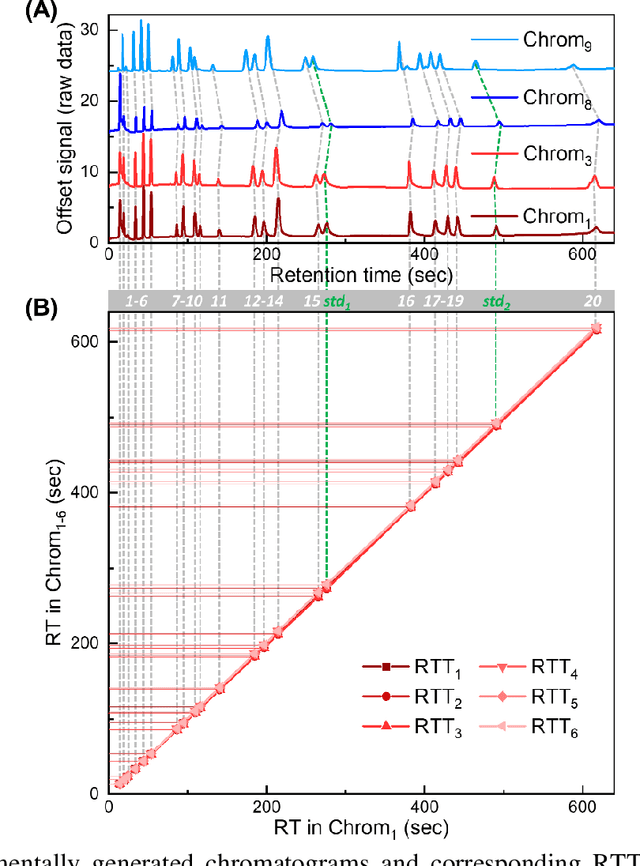
Abstract:Retention time drift caused by fluctuations in physical factors such as temperature ramping rate and carrier gas flow rate is ubiquitous in chromatographic measurements. Proper peak identification and alignment across different chromatograms is critical prior to any subsequent analysis. This work introduces a peak identification method called retention time trajectory (RTT) matching, which uses chromatographic retention times as the only input and identifies peaks associated with any subset of a predefined set of target compounds. RTT matching is also capable of reporting interferents. An RTT is a 2-dimensional (2D) curve formed uniquely by the retention times of the chromatographic peaks. The RTTs obtained from the chromatogram of a test sample and of pre-characterized library are matched and statistically compared. The best matched pair implies identification. Unlike most existing peak alignment methods, no mathematical warping or transformations are involved. Based on the experimentally characterized RTT, an RTT hybridization method is developed to rapidly generate more RTTs without performing actual time-consuming chromatographic measurements. This enables successful identification even for chromatograms with serious retention time drift. Experimentally obtained gas chromatograms and publicly available fruit metabolomics liquid chromatograms are used to generate over two trillions of tests that validate the proposed method, demonstrating real-time peak/interferent identification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge