Wouter van Loon
An introduction to R package `mvs`
Apr 24, 2025



Abstract:In biomedical science, a set of objects or persons can often be described by multiple distinct sets of features obtained from different data sources or modalities (called "multi-view data"). Classical machine learning methods ignore the multi-view structure of such data, limiting model interpretability and performance. The R package `mvs` provides methods that were designed specifically for dealing with multi-view data, based on the multi-view stacking (MVS) framework. MVS is a form of supervised (machine) learning used to train multi-view classification or prediction models. MVS works by training a learning algorithm on each view separately, estimating the predictive power of each view-specific model through cross-validation, and then using another learning algorithm to assign weights to the view-specific models based on their estimated predictions. MVS is a form of ensemble learning, dividing the large multi-view learning problem into smaller sub-problems. Most of these sub-problems can be solved in parallel, making it computationally attractive. Additionally, the number of features of the sub-problems is greatly reduced compared with the full multi-view learning problem. This makes MVS especially useful when the total number of features is larger than the number of observations (i.e., high-dimensional data). MVS can still be applied even if the sub-problems are themselves high-dimensional by adding suitable penalty terms to the learning algorithms. Furthermore, MVS can be used to automatically select the views which are most important for prediction. The R package `mvs` makes fitting MVS models, including such penalty terms, easily and openly accessible. `mvs` allows for the fitting of stacked models with any number of levels, with different penalty terms, different outcome distributions, and provides several options for missing data handling.
Imputation of missing values in multi-view data
Oct 26, 2022



Abstract:When missing values occur in multi-view data, all features in a view are likely to be missing simultaneously. This leads to very large quantities of missing data which, especially when combined with high-dimensionality, makes the application of conditional imputation methods computationally infeasible. We introduce a new meta-learning imputation method based on stacked penalized logistic regression (StaPLR), which performs imputation in a dimension-reduced space. We evaluate the new imputation method with several imputation algorithms using simulations. The results show that meta-level imputation of missing values leads to good results at a much lower computational cost, and makes the use of advanced imputation algorithms such as missForest and predictive mean matching possible in settings where they would otherwise be computationally infeasible.
Analyzing hierarchical multi-view MRI data with StaPLR: An application to Alzheimer's disease classification
Aug 12, 2021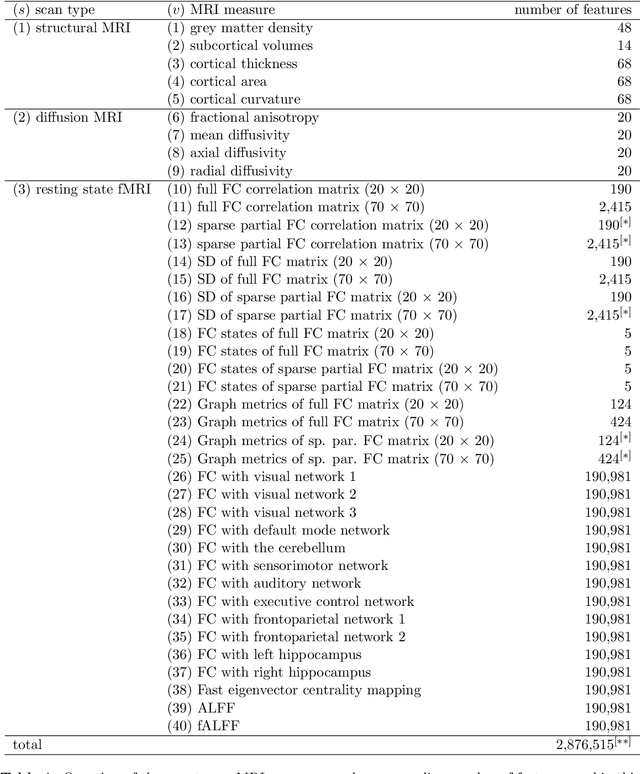
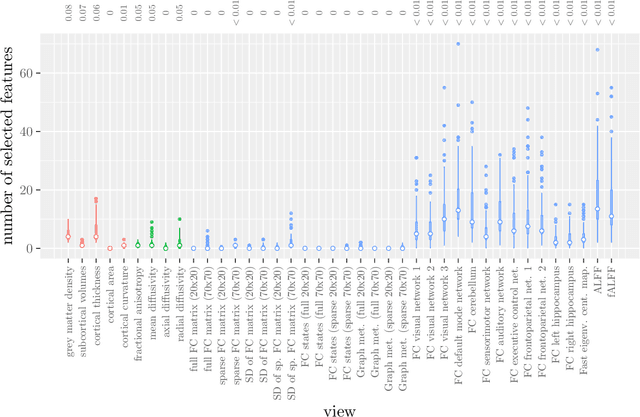
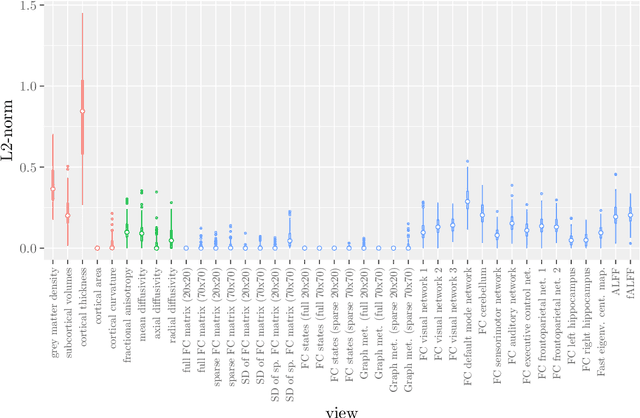
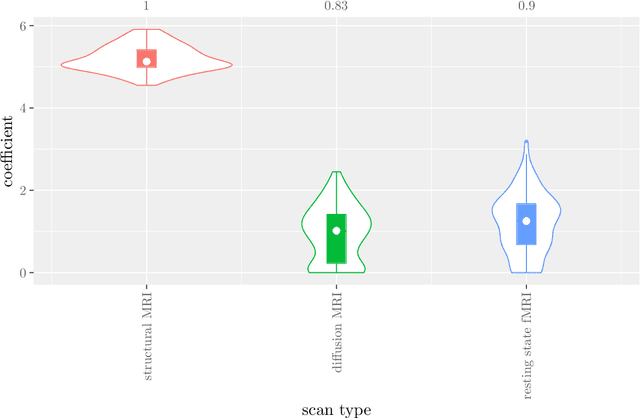
Abstract:Multi-view data refers to a setting where features are divided into feature sets, for example because they correspond to different sources. Stacked penalized logistic regression (StaPLR) is a recently introduced method that can be used for classification and automatically selecting the views that are most important for prediction. We show how this method can easily be extended to a setting where the data has a hierarchical multi-view structure. We apply StaPLR to Alzheimer's disease classification where different MRI measures have been calculated from three scan types: structural MRI, diffusion-weighted MRI, and resting-state fMRI. StaPLR can identify which scan types and which MRI measures are most important for classification, and it outperforms elastic net regression in classification performance.
View selection in multi-view stacking: Choosing the meta-learner
Oct 30, 2020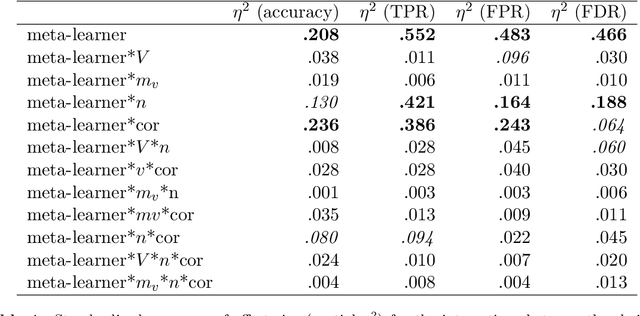
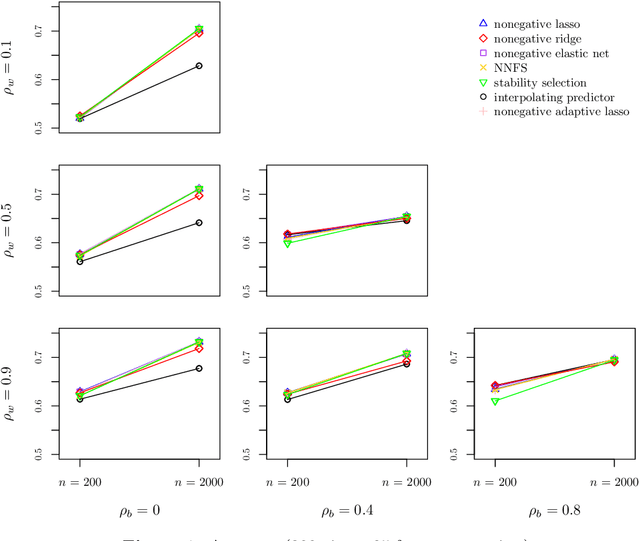
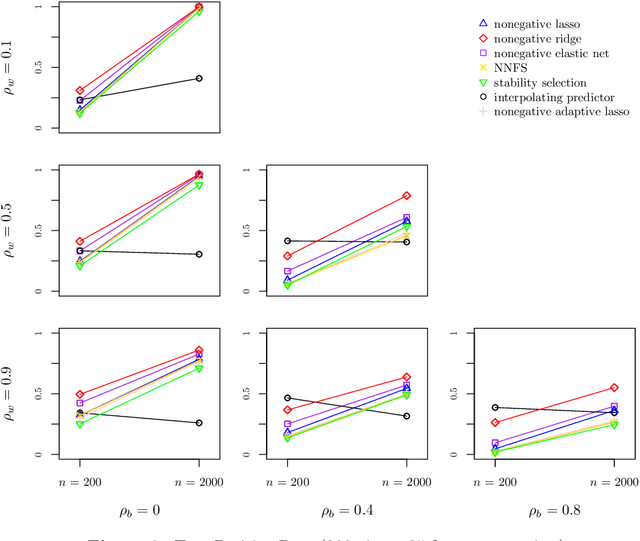
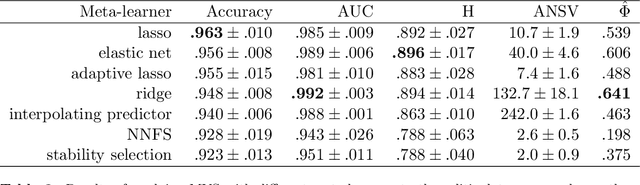
Abstract:Multi-view stacking is a framework for combining information from different views (i.e. different feature sets) describing the same set of objects. In this framework, a base-learner algorithm is trained on each view separately, and their predictions are then combined by a meta-learner algorithm. In a previous study, stacked penalized logistic regression, a special case of multi-view stacking, has been shown to be useful in identifying which views are most important for prediction. In this article we expand this research by considering seven different algorithms to use as the meta-learner, and evaluating their view selection and classification performance in simulations and two applications on real gene-expression data sets. Our results suggest that if both view selection and classification accuracy are important to the research at hand, then the nonnegative lasso, nonnegative adaptive lasso and nonnegative elastic net are suitable meta-learners. Exactly which among these three is to be preferred depends on the research context. The remaining four meta-learners, namely nonnegative ridge regression, nonnegative forward selection, stability selection and the interpolating predictor, show little advantages in order to be preferred over the other three.
Stacked Penalized Logistic Regression for Selecting Views in Multi-View Learning
Nov 06, 2018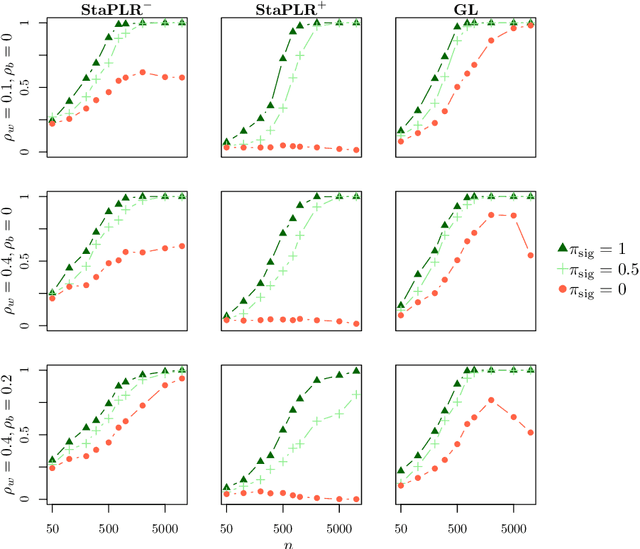
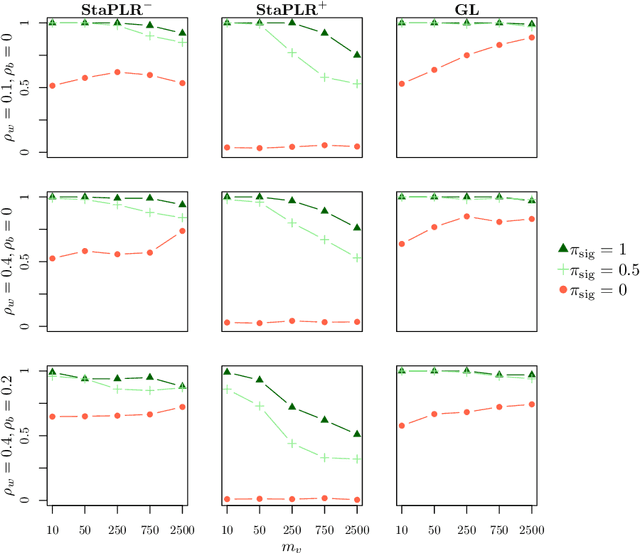
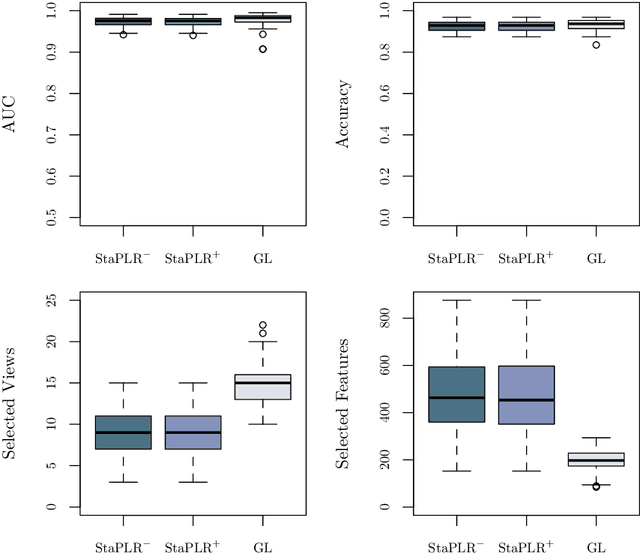
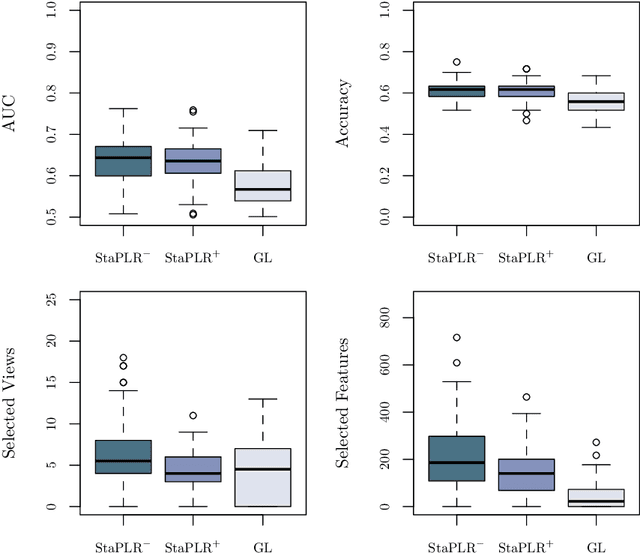
Abstract:In multi-view learning, features are organized into multiple sets called views. Multi-view stacking (MVS) is an ensemble learning framework which learns a prediction function from each view separately, and then learns a meta-function which optimally combines the view-specific predictions. In case studies, MVS has been shown to increase prediction accuracy. However, the framework can also be used for selecting a subset of important views. We propose a method for selecting views based on MVS, which we call stacked penalized logistic regression (StaPLR). Compared to existing view-selection methods like the group lasso, StaPLR can make use of faster optimization algorithms and is easily parallelized. We show that nonnegativity constraints on the parameters of the function which combines the views are important for preventing unimportant views from entering the model. We investigate the view selection and classification performance of StaPLR and the group lasso through simulations, and consider two real data examples. We observe that StaPLR is less likely to select irrelevant views, leading to models that are sparser at the view level, but which have comparable or increased predictive performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge