Toby D. Hocking
Fused Lasso Improves Accuracy of Co-occurrence Network Inference in Grouped Samples
Sep 11, 2025Abstract:Co-occurrence network inference algorithms have significantly advanced our understanding of microbiome communities. However, these algorithms typically analyze microbial associations within samples collected from a single environmental niche, often capturing only static snapshots rather than dynamic microbial processes. Previous studies have commonly grouped samples from different environmental niches together without fully considering how microbial communities adapt their associations when faced with varying ecological conditions. Our study addresses this limitation by explicitly investigating both spatial and temporal dynamics of microbial communities. We analyzed publicly available microbiome abundance data across multiple locations and time points, to evaluate algorithm performance in predicting microbial associations using our proposed Same-All Cross-validation (SAC) framework. SAC evaluates algorithms in two distinct scenarios: training and testing within the same environmental niche (Same), and training and testing on combined data from multiple environmental niches (All). To overcome the limitations of conventional algorithms, we propose fuser, an algorithm that, while not entirely new in machine learning, is novel for microbiome community network inference. It retains subsample-specific signals while simultaneously sharing relevant information across environments during training. Unlike standard approaches that infer a single generalized network from combined data, fuser generates distinct, environment-specific predictive networks. Our results demonstrate that fuser achieves comparable predictive performance to existing algorithms such as glmnet when evaluated within homogeneous environments (Same), and notably reduces test error compared to baseline algorithms in cross-environment (All) scenarios.
A Log-linear Gradient Descent Algorithm for Unbalanced Binary Classification using the All Pairs Squared Hinge Loss
Feb 21, 2023Abstract:Receiver Operating Characteristic (ROC) curves are plots of true positive rate versus false positive rate which are used to evaluate binary classification algorithms. Because the Area Under the Curve (AUC) is a constant function of the predicted values, learning algorithms instead optimize convex relaxations which involve a sum over all pairs of labeled positive and negative examples. Naive learning algorithms compute the gradient in quadratic time, which is too slow for learning using large batch sizes. We propose a new functional representation of the square loss and squared hinge loss, which results in algorithms that compute the gradient in either linear or log-linear time, and makes it possible to use gradient descent learning with large batch sizes. In our empirical study of supervised binary classification problems, we show that our new algorithm can achieve higher test AUC values on imbalanced data sets than previous algorithms, and make use of larger batch sizes than were previously feasible.
Functional Labeled Optimal Partitioning
Oct 05, 2022
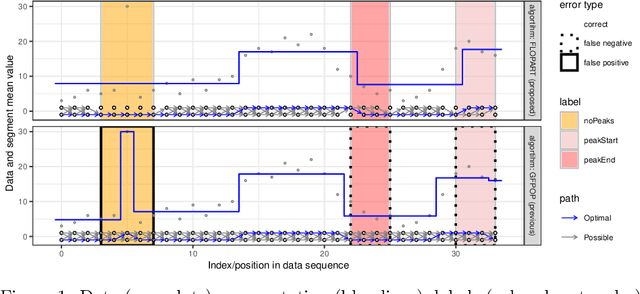
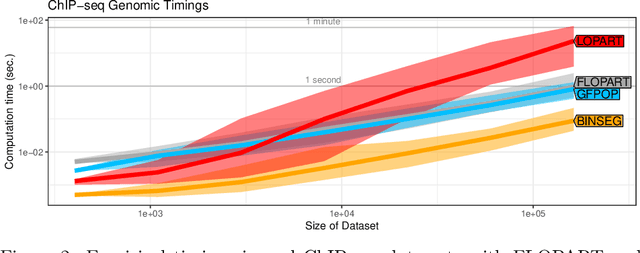
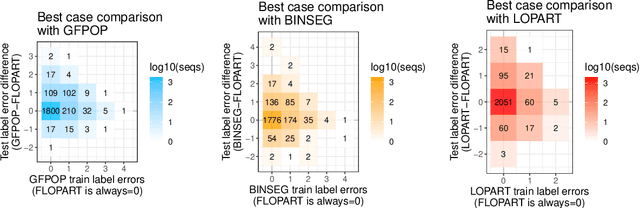
Abstract:Peak detection is a problem in sequential data analysis that involves differentiating regions with higher counts (peaks) from regions with lower counts (background noise). It is crucial to correctly predict areas that deviate from the background noise, in both the train and test sets of labels. Dynamic programming changepoint algorithms have been proposed to solve the peak detection problem by constraining the mean to alternatively increase and then decrease. The current constrained changepoint algorithms only create predictions on the test set, while completely ignoring the train set. Changepoint algorithms that are both accurate when fitting the train set, and make predictions on the test set, have been proposed but not in the context of peak detection models. We propose to resolve these issues by creating a new dynamic programming algorithm, FLOPART, that has zero train label errors, and is able to provide highly accurate predictions on the test set. We provide an empirical analysis that shows FLOPART has a similar time complexity while being more accurate than the existing algorithms in terms of train and test label errors.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge