Temiloluwa Prioleau
Glucose-ML: A collection of longitudinal diabetes datasets for development of robust AI solutions
Jul 18, 2025Abstract:Artificial intelligence (AI) algorithms are a critical part of state-of-the-art digital health technology for diabetes management. Yet, access to large high-quality datasets is creating barriers that impede development of robust AI solutions. To accelerate development of transparent, reproducible, and robust AI solutions, we present Glucose-ML, a collection of 10 publicly available diabetes datasets, released within the last 7 years (i.e., 2018 - 2025). The Glucose-ML collection comprises over 300,000 days of continuous glucose monitor (CGM) data with a total of 38 million glucose samples collected from 2500+ people across 4 countries. Participants include persons living with type 1 diabetes, type 2 diabetes, prediabetes, and no diabetes. To support researchers and innovators with using this rich collection of diabetes datasets, we present a comparative analysis to guide algorithm developers with data selection. Additionally, we conduct a case study for the task of blood glucose prediction - one of the most common AI tasks within the field. Through this case study, we provide a benchmark for short-term blood glucose prediction across all 10 publicly available diabetes datasets within the Glucose-ML collection. We show that the same algorithm can have significantly different prediction results when developed/evaluated with different datasets. Findings from this study are then used to inform recommendations for developing robust AI solutions within the diabetes or broader health domain. We provide direct links to each longitudinal diabetes dataset in the Glucose-ML collection and openly provide our code.
DiaTrend: A dataset from advanced diabetes technology to enable development of novel analytic solutions
Apr 04, 2023Abstract:Objective digital data is scarce yet needed in many domains to enable research that can transform the standard of healthcare. While data from consumer-grade wearables and smartphones is more accessible, there is critical need for similar data from clinical-grade devices used by patients with a diagnosed condition. The prevalence of wearable medical devices in the diabetes domain sets the stage for unique research and development within this field and beyond. However, the scarcity of open-source datasets presents a major barrier to progress. To facilitate broader research on diabetes-relevant problems and accelerate development of robust computational solutions, we provide the DiaTrend dataset. The DiaTrend dataset is composed of intensive longitudinal data from wearable medical devices, including a total of 27,561 days of continuous glucose monitor data and 8,220 days of insulin pump data from 54 patients with diabetes. This dataset is useful for developing novel analytic solutions that can reduce the disease burden for people living with diabetes and increase knowledge on chronic condition management in outpatient settings.
Feature Selection for Multivariate Time Series via Network Pruning
Feb 11, 2021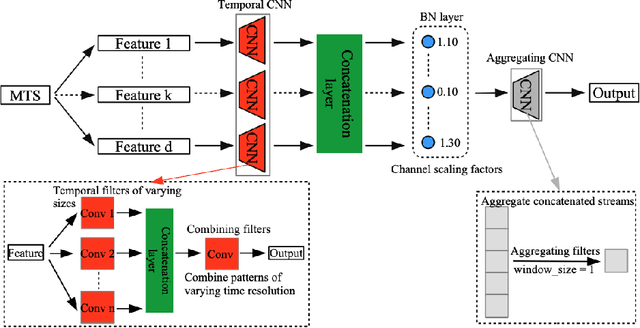
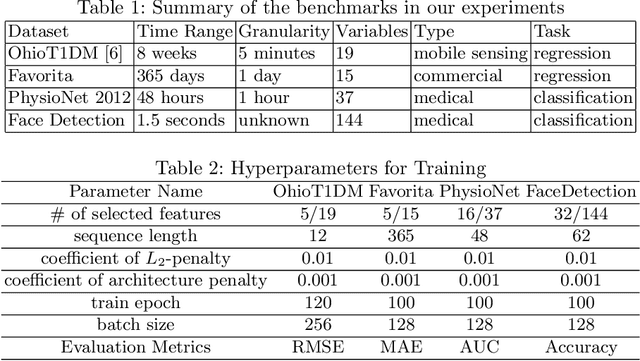

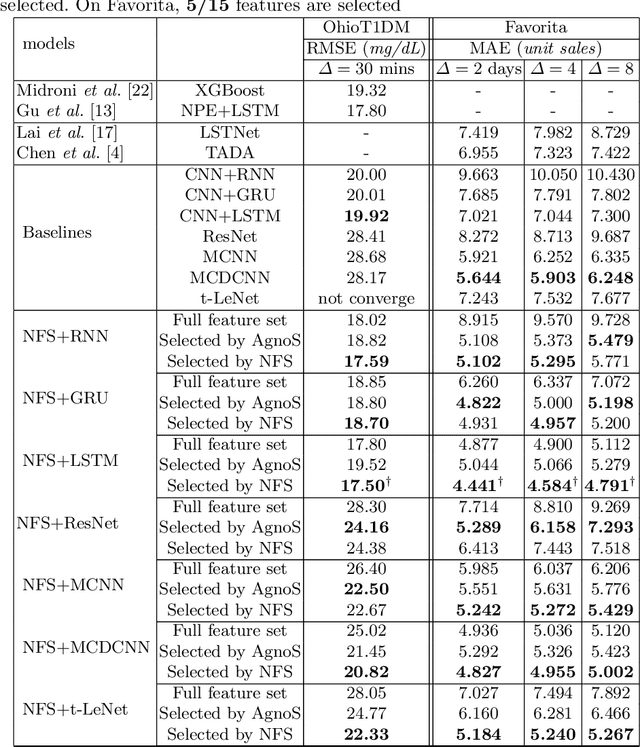
Abstract:In recent years, there has been an ever increasing amount of multivariate time series (MTS) data in various domains, typically generated by a large family of sensors such as wearable devices. This has led to the development of novel learning methods on MTS data, with deep learning models dominating the most recent advancements. Prior literature has primarily focused on designing new network architectures for modeling temporal dependencies within MTS. However, a less studied challenge is associated with high dimensionality of MTS data. In this paper, we propose a novel neural component, namely Neural Feature Se-lector (NFS), as an end-2-end solution for feature selection in MTS data. Specifically, NFS is based on decomposed convolution design and includes two modules: firstly each feature stream within MTS is processed by a temporal CNN independently; then an aggregating CNN combines the processed streams to produce input for other downstream networks. We evaluated the proposed NFS model on four real-world MTS datasets and found that it achieves comparable results with state-of-the-art methods while providing the benefit of feature selection. Our paper also highlights the robustness and effectiveness of feature selection with NFS compared to using recent autoencoder-based methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge