Sulaiman Vesal
SkinNet: A Deep Learning Framework for Skin Lesion Segmentation
Jun 25, 2018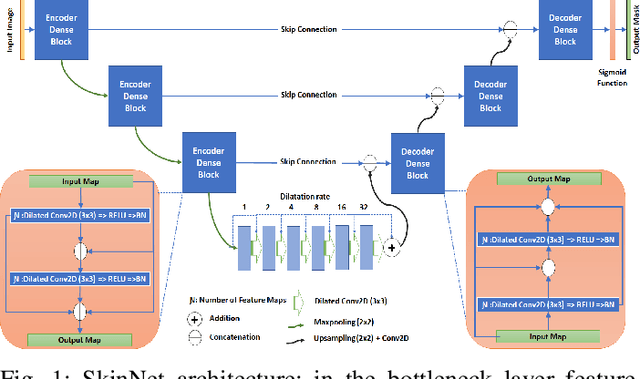
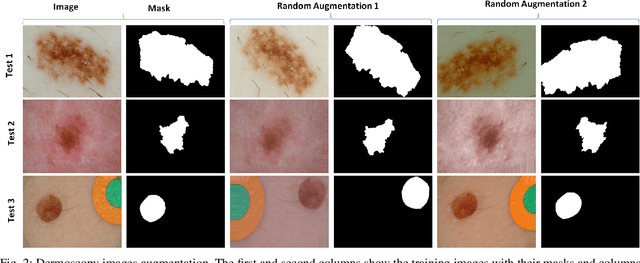
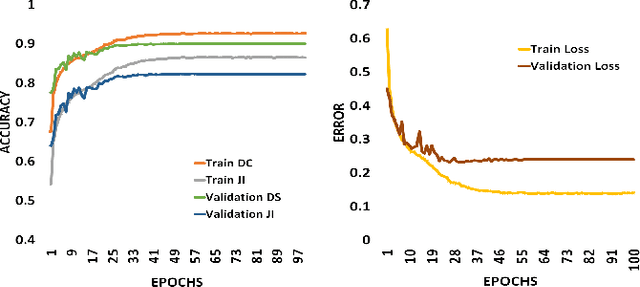
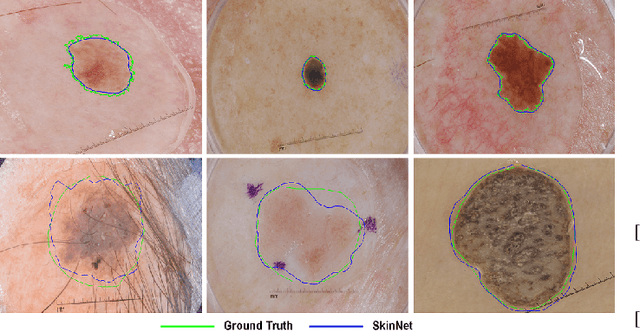
Abstract:There has been a steady increase in the incidence of skin cancer worldwide, with a high rate of mortality. Early detection and segmentation of skin lesions are crucial for timely diagnosis and treatment, necessary to improve the survival rate of patients. However, skin lesion segmentation is a challenging task due to the low contrast of lesions and their high similarity in terms of appearance, to healthy tissue. This underlines the need for an accurate and automatic approach for skin lesion segmentation. To tackle this issue, we propose a convolutional neural network (CNN) called SkinNet. The proposed CNN is a modified version of U-Net. We compared the performance of our approach with other state-of-the-art techniques, using the ISBI 2017 challenge dataset. Our approach outperformed the others in terms of the Dice coefficient, Jaccard index and sensitivity, evaluated on the held-out challenge test data set, across 5-fold cross validation experiments. SkinNet achieved an average value of 85.10, 76.67 and 93.0%, for the DC, JI, and SE, respectively.
Hyper-Hue and EMAP on Hyperspectral Images for Supervised Layer Decomposition of Old Master Drawings
May 28, 2018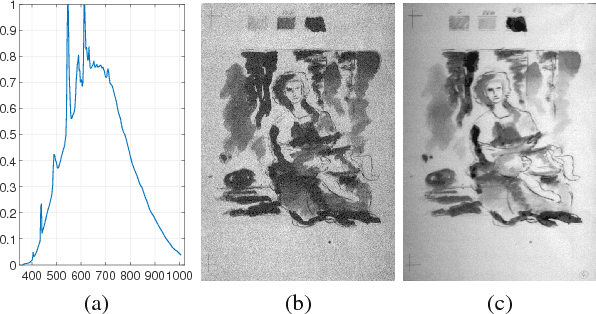
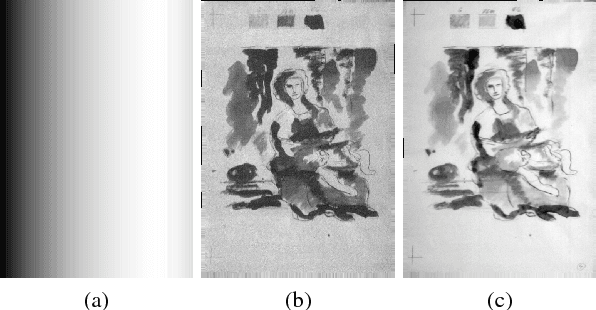


Abstract:Old master drawings were mostly created step by step in several layers using different materials. To art historians and restorers, examination of these layers brings various insights into the artistic work process and helps to answer questions about the object, its attribution and its authenticity. However, these layers typically overlap and are oftentimes difficult to differentiate with the unaided eye. For example, a common layer combination is red chalk under ink. In this work, we propose an image processing pipeline that operates on hyperspectral images to separate such layers. Using this pipeline, we show that hyperspectral images enable better layer separation than RGB images, and that spectral focus stacking aids the layer separation. In particular, we propose to use two descriptors in hyperspectral historical document analysis, namely hyper-hue and extended multi-attribute profile (EMAP). Our comparative results with other features underline the efficacy of the three proposed improvements.
Classification of breast cancer histology images using transfer learning
Feb 26, 2018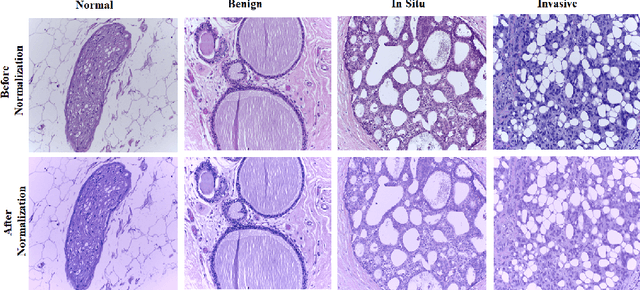
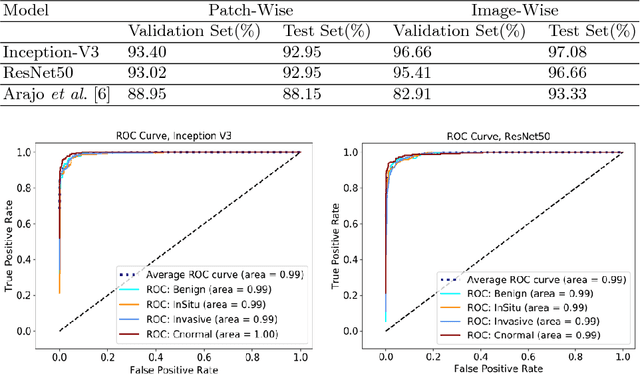
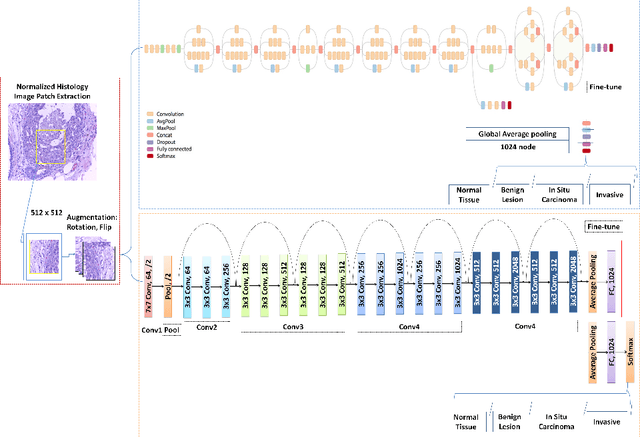
Abstract:Breast cancer is one of the leading causes of mortality in women. Early detection and treatment are imperative for improving survival rates, which have steadily increased in recent years as a result of more sophisticated computer-aided-diagnosis (CAD) systems. A critical component of breast cancer diagnosis relies on histopathology, a laborious and highly subjective process. Consequently, CAD systems are essential to reduce inter-rater variability and supplement the analyses conducted by specialists. In this paper, a transfer-learning based approach is proposed, for the task of breast histology image classification into four tissue sub-types, namely, normal, benign, \textit{in situ} carcinoma and invasive carcinoma. The histology images, provided as part of the BACH 2018 grand challenge, were first normalized to correct for color variations resulting from inconsistencies during slide preparation. Subsequently, image patches were extracted and used to fine-tune Google`s Inception-V3 and ResNet50 convolutional neural networks (CNNs), both pre-trained on the ImageNet database, enabling them to learn domain-specific features, necessary to classify the histology images. The ResNet50 network (based on residual learning) achieved a test classification accuracy of 97.50% for four classes, outperforming the Inception-V3 network which achieved an accuracy of 91.25%.
Comparative Analysis of Unsupervised Algorithms for Breast MRI Lesion Segmentation
Feb 23, 2018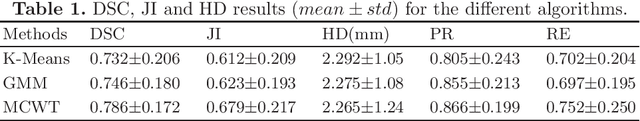
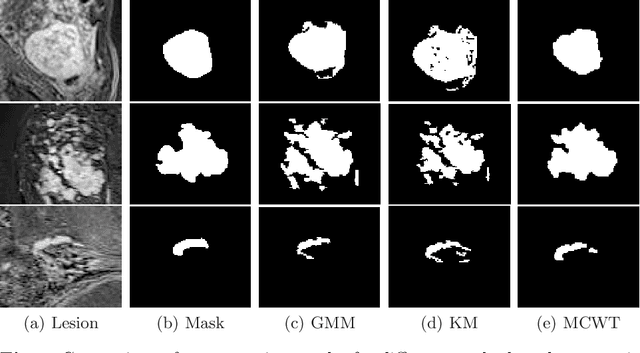
Abstract:Accurate segmentation of breast lesions is a crucial step in evaluating the characteristics of tumors. However, this is a challenging task, since breast lesions have sophisticated shape, topological structure, and variation in the intensity distribution. In this paper, we evaluated the performance of three unsupervised algorithms for the task of breast Magnetic Resonance (MRI) lesion segmentation, namely, Gaussian Mixture Model clustering, K-means clustering and a marker-controlled Watershed transformation based method. All methods were applied on breast MRI slices following selection of regions of interest (ROIs) by an expert radiologist and evaluated on 106 subjects' images, which include 59 malignant and 47 benign lesions. Segmentation accuracy was evaluated by comparing our results with ground truth masks, using the Dice similarity coefficient (DSC), Jaccard index (JI), Hausdorff distance and precision-recall metrics. The results indicate that the marker-controlled Watershed transformation outperformed all other algorithms investigated.
Semi-Automatic Algorithm for Breast MRI Lesion Segmentation Using Marker-Controlled Watershed Transformation
Dec 14, 2017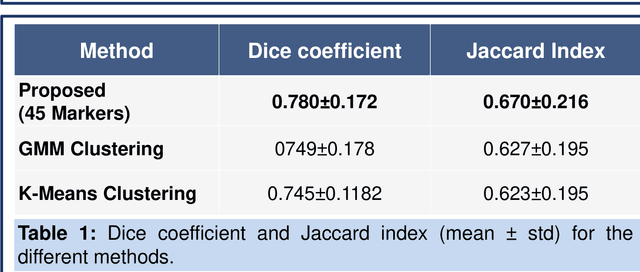
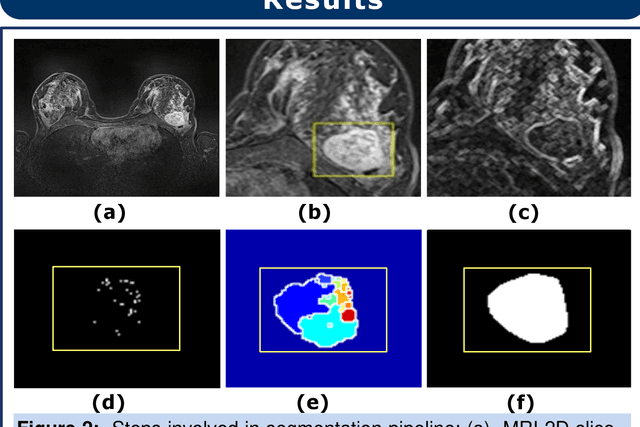
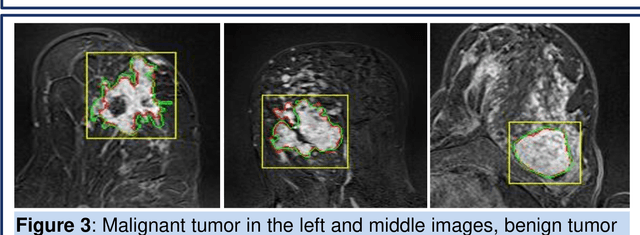
Abstract:Magnetic resonance imaging (MRI) is an effective imaging modality for identifying and localizing breast lesions in women. Accurate and precise lesion segmentation using a computer-aided-diagnosis (CAD) system, is a crucial step in evaluating tumor volume and in the quantification of tumor characteristics. However, this is a challenging task, since breast lesions have sophisticated shape, topological structure, and high variance in their intensity distribution across patients. In this paper, we propose a novel marker-controlled watershed transformation-based approach, which uses the brightest pixels in a region of interest (determined by experts) as markers to overcome this challenge, and accurately segment lesions in breast MRI. The proposed approach was evaluated on 106 lesions, which includes 64 malignant and 42 benign cases. Segmentation results were quantified by comparison with ground truth labels, using the Dice similarity coefficient (DSC) and Jaccard index (JI) metrics. The proposed method achieved an average Dice coefficient of 0.7808$\pm$0.1729 and Jaccard index of 0.6704$\pm$0.2167. These results illustrate that the proposed method shows promise for future work related to the segmentation and classification of benign and malignant breast lesions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge