Stephen D. H. Hsu
Embedded AI Companion System on Edge Devices
Jan 13, 2026Abstract:Computational resource constraints on edge devices make it difficult to develop a fully embedded AI companion system with a satisfactory user experience. AI companion and memory systems detailed in existing literature cannot be directly used in such an environment due to lack of compute resources and latency concerns. In this paper, we propose a memory paradigm that alternates between active and inactive phases: during phases of user activity, the system performs low-latency, real-time dialog using lightweight retrieval over existing memories and context; whereas during phases of user inactivity, it conducts more computationally intensive extraction, consolidation, and maintenance of memories across full conversation sessions. This design minimizes latency while maintaining long-term personalization under the tight constraints of embedded hardware. We also introduce an AI Companion benchmark designed to holistically evaluate the AI Companion across both its conversational quality and memory capabilities. In our experiments, we found that our system (using a very weak model: Qwen2.5-7B-Instruct quantized int4) outperforms the equivalent raw LLM without memory across most metrics, and performs comparably to GPT-3.5 with 16k context window.
Accurate Genomic Prediction Of Human Height
Sep 19, 2017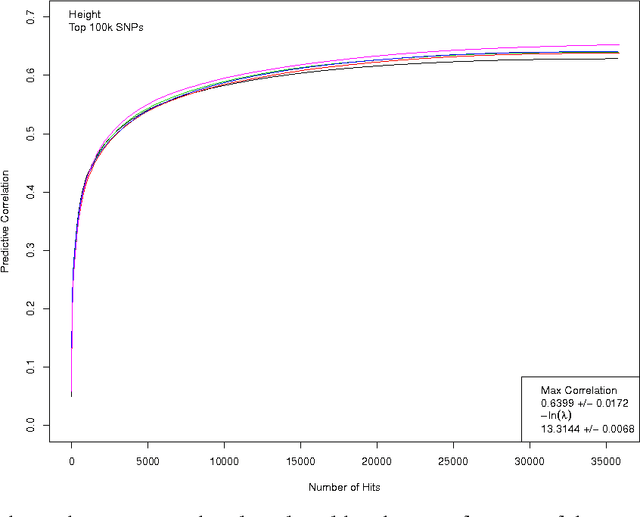
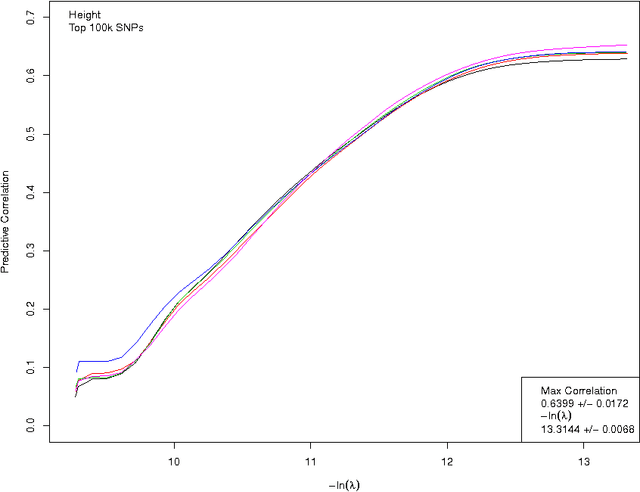
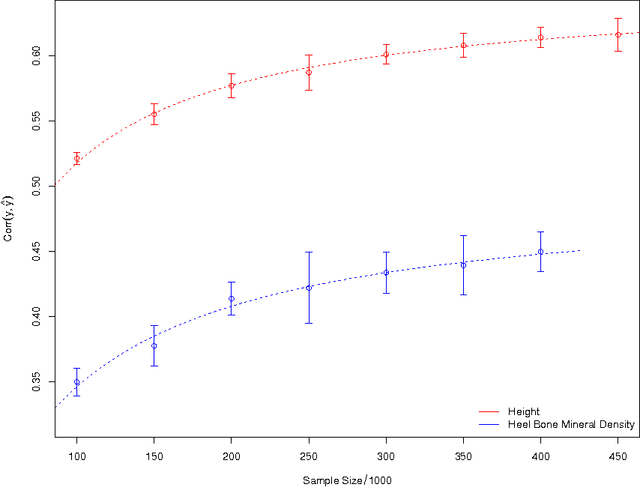
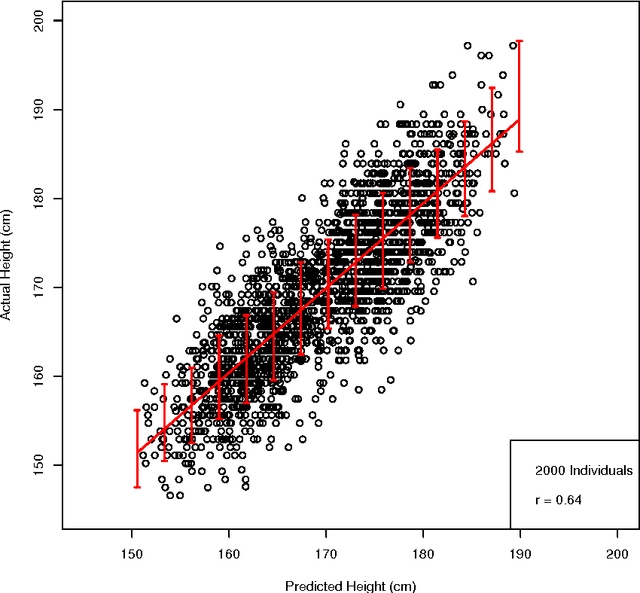
Abstract:We construct genomic predictors for heritable and extremely complex human quantitative traits (height, heel bone density, and educational attainment) using modern methods in high dimensional statistics (i.e., machine learning). Replication tests show that these predictors capture, respectively, $\sim$40, 20, and 9 percent of total variance for the three traits. For example, predicted heights correlate $\sim$0.65 with actual height; actual heights of most individuals in validation samples are within a few cm of the prediction. The variance captured for height is comparable to the estimated SNP heritability from GCTA (GREML) analysis, and seems to be close to its asymptotic value (i.e., as sample size goes to infinity), suggesting that we have captured most of the heritability for the SNPs used. Thus, our results resolve the common SNP portion of the "missing heritability" problem -- i.e., the gap between prediction R-squared and SNP heritability. The $\sim$20k activated SNPs in our height predictor reveal the genetic architecture of human height, at least for common SNPs. Our primary dataset is the UK Biobank cohort, comprised of almost 500k individual genotypes with multiple phenotypes. We also use other datasets and SNPs found in earlier GWAS for out-of-sample validation of our results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge