Shadi Albarqouni
University Hospital Bonn, Venusberg-Campus 1, D-53127, Bonn, Germany, Helmholtz Munich, Ingolstädter Landstraße 1, D-85764, Neuherberg, Germany, Technical University of Munich, Boltzmannstr. 3, D-85748 Garching, Germany
MelanoGANs: High Resolution Skin Lesion Synthesis with GANs
Apr 12, 2018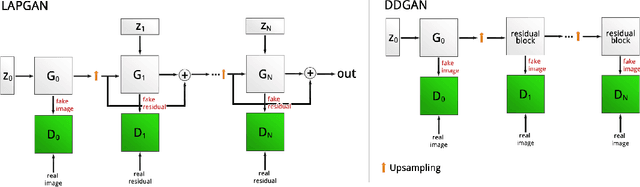
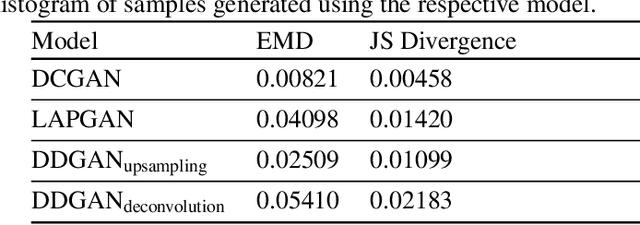
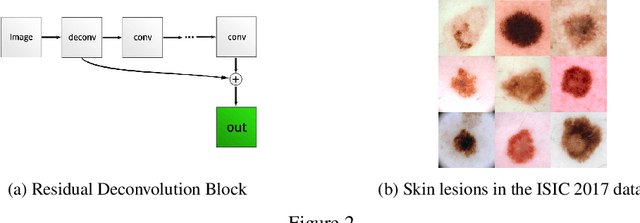

Abstract:Generative Adversarial Networks (GANs) have been successfully used to synthesize realistically looking images of faces, scenery and even medical images. Unfortunately, they usually require large training datasets, which are often scarce in the medical field, and to the best of our knowledge GANs have been only applied for medical image synthesis at fairly low resolution. However, many state-of-the-art machine learning models operate on high resolution data as such data carries indispensable, valuable information. In this work, we try to generate realistically looking high resolution images of skin lesions with GANs, using only a small training dataset of 2000 samples. The nature of the data allows us to do a direct comparison between the image statistics of the generated samples and the real dataset. We both quantitatively and qualitatively compare state-of-the-art GAN architectures such as DCGAN and LAPGAN against a modification of the latter for the task of image generation at a resolution of 256x256px. Our investigation shows that we can approximate the real data distribution with all of the models, but we notice major differences when visually rating sample realism, diversity and artifacts. In a set of use-case experiments on skin lesion classification, we further show that we can successfully tackle the problem of heavy class imbalance with the help of synthesized high resolution melanoma samples.
StainGAN: Stain Style Transfer for Digital Histological Images
Apr 04, 2018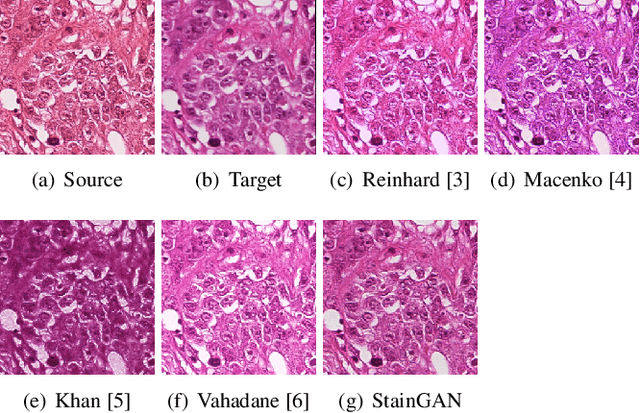

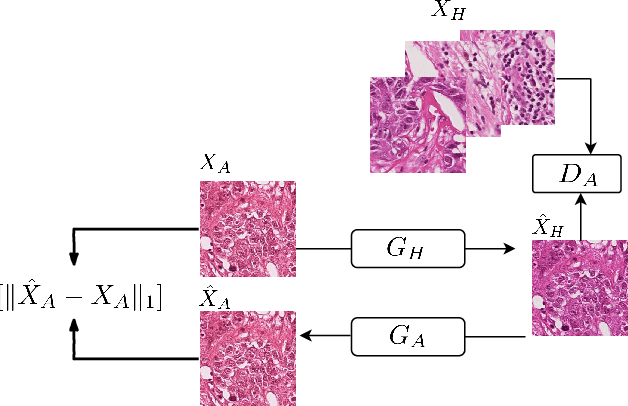
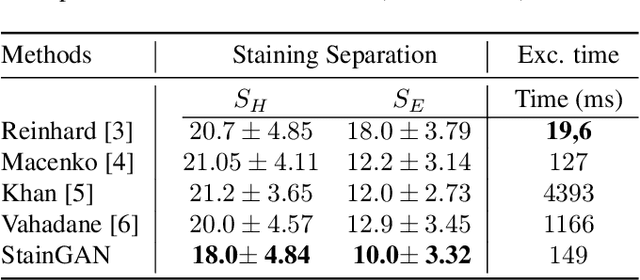
Abstract:Digitized Histological diagnosis is in increasing demand. However, color variations due to various factors are imposing obstacles to the diagnosis process. The problem of stain color variations is a well-defined problem with many proposed solutions. Most of these solutions are highly dependent on a reference template slide. We propose a deep-learning solution inspired by CycleGANs that is trained end-to-end, eliminating the need for an expert to pick a representative reference slide. Our approach showed superior results quantitatively and qualitatively against the state of the art methods (10% improvement visually using SSIM). We further validated our method on a clinical use-case, namely Breast Cancer tumor classification, showing 12% increase in AUC. The code will be made publicly available.
Semi-Supervised Deep Learning for Fully Convolutional Networks
Jul 25, 2017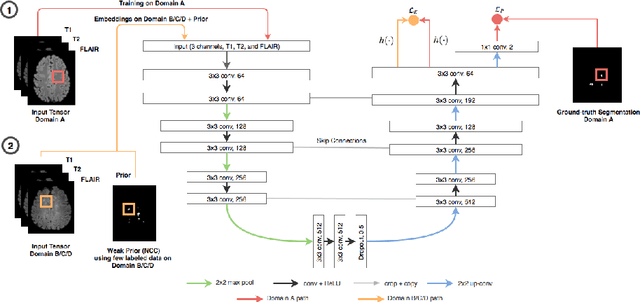

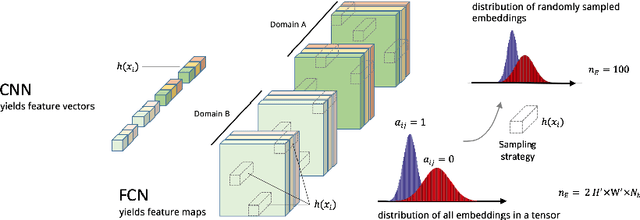
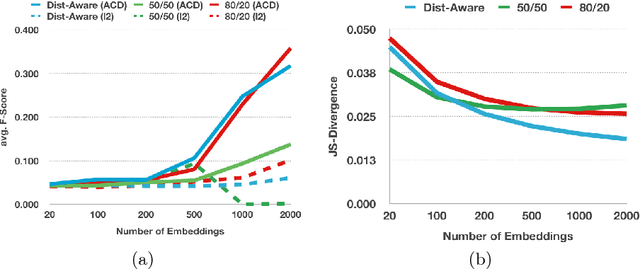
Abstract:Deep learning usually requires large amounts of labeled training data, but annotating data is costly and tedious. The framework of semi-supervised learning provides the means to use both labeled data and arbitrary amounts of unlabeled data for training. Recently, semi-supervised deep learning has been intensively studied for standard CNN architectures. However, Fully Convolutional Networks (FCNs) set the state-of-the-art for many image segmentation tasks. To the best of our knowledge, there is no existing semi-supervised learning method for such FCNs yet. We lift the concept of auxiliary manifold embedding for semi-supervised learning to FCNs with the help of Random Feature Embedding. In our experiments on the challenging task of MS Lesion Segmentation, we leverage the proposed framework for the purpose of domain adaptation and report substantial improvements over the baseline model.
X-ray In-Depth Decomposition: Revealing The Latent Structures
Mar 22, 2017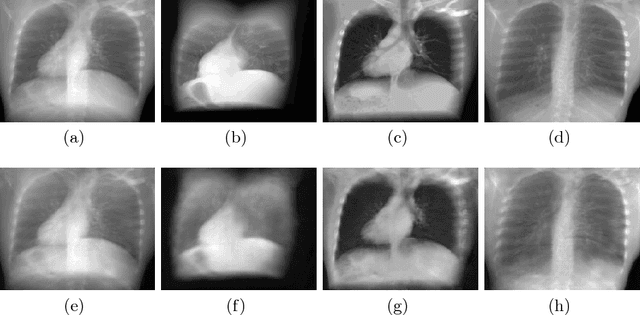
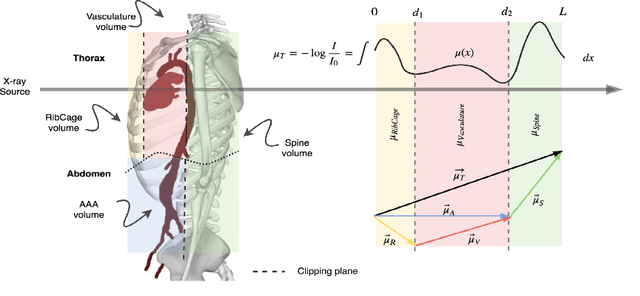
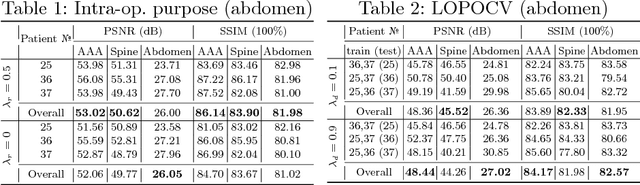
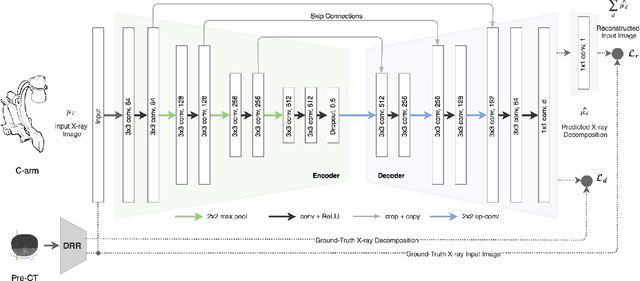
Abstract:X-ray radiography is the most readily available imaging modality and has a broad range of applications that spans from diagnosis to intra-operative guidance in cardiac, orthopedics, and trauma procedures. Proper interpretation of the hidden and obscured anatomy in X-ray images remains a challenge and often requires high radiation dose and imaging from several perspectives. In this work, we aim at decomposing the conventional X-ray image into d X-ray components of independent, non-overlapped, clipped sub-volumes using deep learning approach. Despite the challenging aspects of modeling such a highly ill-posed problem, exciting and encouraging results are obtained paving the path for further contributions in this direction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge