Sean D. Stocker
Quantifying Local Strain Field and Deformation in Active Contraction of Bladder Using a Pretrained Transformer Model: A Speckle-Free Approach
Jan 04, 2026Abstract:Accurate quantification of local strain fields during bladder contraction is essential for understanding the biomechanics of bladder micturition, in both health and disease. Conventional digital image correlation (DIC) methods have been successfully applied to various biological tissues; however, this approach requires artificial speckling, which can alter both passive and active properties of the tissue. In this study, we introduce a speckle-free framework for quantifying local strain fields using a state-of-the-art, zero-shot transformer model, CoTracker3. We utilized a custom-designed, portable isotonic biaxial apparatus compatible with multiphoton microscopy (MPM) to demonstrate this approach, successfully tracking natural bladder lumen textures without artificial markers. Benchmark tests validated the method's high pixel accuracy and low strain errors. Our framework effectively captured heterogeneous deformation patterns, despite complex folding and buckling, which conventional DIC often fails to track. Application to in vitro active bladder contractions in four rat specimens (n=4) revealed statistically significant anisotropy (p<0.01), with higher contraction longitudinally compared to circumferentially. Multiphoton microscopy further illustrated and confirmed heterogeneous morphological changes, such as large fold formation during active contraction. This non-invasive approach eliminates speckle-induced artifacts, enabling more physiologically relevant measurements, and has broad applicability for material testing of other biological and engineered systems.
Quantifying Smooth Muscles Regional Organization in the Rat Bladder Using Immunohistochemistry, Multiphoton Microscopy and Machine Learning
May 08, 2024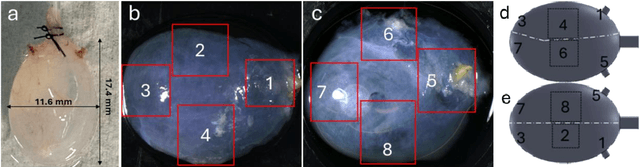
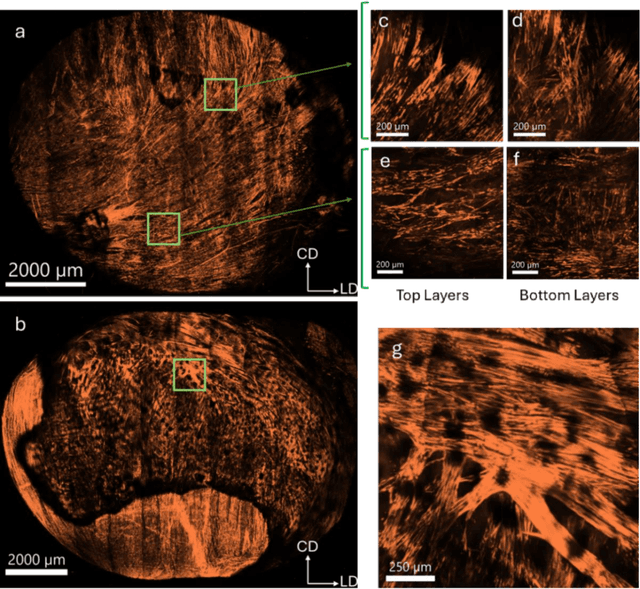

Abstract:The smooth muscle bundles (SMBs) in the bladder act as contractile elements which enable the bladder to void effectively. In contrast to skeletal muscles, these bundles are not highly aligned, rather they are oriented more heterogeneously throughout the bladder wall. In this work, for the first time, this regional orientation of the SMBs is quantified across the whole bladder, without the need for optical clearing or cryosectioning. Immunohistochemistry staining was utilized to visualize smooth muscle cell actin in multiphoton microscopy (MPM) images of bladder smooth muscle bundles (SMBs). Feature vectors for each pixel were generated using a range of filters, including Gaussian blur, Gaussian gradient magnitude, Laplacian of Gaussian, Hessian eigenvalues, structure tensor eigenvalues, Gabor, and Sobel gradients. A Random Forest classifier was subsequently trained to automate the segmentation of SMBs in the MPM images. Finally, the orientation of SMBs in each bladder region was quantified using the CT-FIRE package. This information is essential for biomechanical models of the bladder that include contractile elements.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge