Sadasivan Puthusserypady
Leveraging Self-Supervised Learning Methods for Remote Screening of Subjects with Paroxysmal Atrial Fibrillation
Mar 04, 2025


Abstract:The integration of Artificial Intelligence (AI) into clinical research has great potential to reveal patterns that are difficult for humans to detect, creating impactful connections between inputs and clinical outcomes. However, these methods often require large amounts of labeled data, which can be difficult to obtain in healthcare due to strict privacy laws and the need for experts to annotate data. This requirement creates a bottleneck when investigating unexplored clinical questions. This study explores the application of Self-Supervised Learning (SSL) as a way to obtain preliminary results from clinical studies with limited sized cohorts. To assess our approach, we focus on an underexplored clinical task: screening subjects for Paroxysmal Atrial Fibrillation (P-AF) using remote monitoring, single-lead ECG signals captured during normal sinus rhythm. We evaluate state-of-the-art SSL methods alongside supervised learning approaches, where SSL outperforms supervised learning in this task of interest. More importantly, it prevents misleading conclusions that may arise from poor performance in the latter paradigm when dealing with limited cohort settings.
CuPID: Leveraging Masked Single-Lead ECG Modelling for Enhancing the Representations
Feb 28, 2025



Abstract:Wearable sensing devices, such as Electrocardiogram (ECG) heart-rate monitors, will play a crucial role in the future of digital health. This continuous monitoring leads to massive unlabeled data, incentivizing the development of unsupervised learning frameworks. While Masked Data Modelling (MDM) techniques have enjoyed wide use, their direct application to single-lead ECG data is suboptimal due to the decoder's difficulty handling irregular heartbeat intervals when no contextual information is provided. In this paper, we present Cueing the Predictor Increments the Detailing (CuPID), a novel MDM method tailored to single-lead ECGs. CuPID enhances existing MDM techniques by cueing spectrogram-derived context to the decoder, thus incentivizing the encoder to produce more detailed representations. This has a significant impact on the encoder's performance across a wide range of different configurations, leading CuPID to outperform state-of-the-art methods in a variety of downstream tasks.
Parallel-Learning of Invariant and Tempo-variant Attributes of Single-Lead Cardiac Signals: PLITA
Feb 28, 2025Abstract:Wearable sensing devices, such as Holter monitors, will play a crucial role in the future of digital health. Unsupervised learning frameworks such as Self-Supervised Learning (SSL) are essential to map these single-lead electrocardiogram (ECG) signals with their anticipated clinical outcomes. These signals are characterized by a tempo-variant component whose patterns evolve through the recording and an invariant component with patterns that remain unchanged. However, existing SSL methods only drive the model to encode the invariant attributes, leading the model to neglect tempo-variant information which reflects subject-state changes through time. In this paper, we present Parallel-Learning of Invariant and Tempo-variant Attributes (PLITA), a novel SSL method designed for capturing both invariant and tempo-variant ECG attributes. The latter are captured by mandating closer representations in space for closer inputs on time. We evaluate both the capability of the method to learn the attributes of these two distinct kinds, as well as PLITA's performance compared to existing SSL methods for ECG analysis. PLITA performs significantly better in the set-ups where tempo-variant attributes play a major role.
Contrastive Learning Is Not Optimal for Quasiperiodic Time Series
Jul 24, 2024



Abstract:Despite recent advancements in Self-Supervised Learning (SSL) for time series analysis, a noticeable gap persists between the anticipated achievements and actual performance. While these methods have demonstrated formidable generalization capabilities with minimal labels in various domains, their effectiveness in distinguishing between different classes based on a limited number of annotated records is notably lacking. Our hypothesis attributes this bottleneck to the prevalent use of Contrastive Learning, a shared training objective in previous state-of-the-art (SOTA) methods. By mandating distinctiveness between representations for negative pairs drawn from separate records, this approach compels the model to encode unique record-based patterns but simultaneously neglects changes occurring across the entire record. To overcome this challenge, we introduce Distilled Embedding for Almost-Periodic Time Series (DEAPS) in this paper, offering a non-contrastive method tailored for quasiperiodic time series, such as electrocardiogram (ECG) data. By avoiding the use of negative pairs, we not only mitigate the model's blindness to temporal changes but also enable the integration of a "Gradual Loss (Lgra)" function. This function guides the model to effectively capture dynamic patterns evolving throughout the record. The outcomes are promising, as DEAPS demonstrates a notable improvement of +10% over existing SOTA methods when just a few annotated records are presented to fit a Machine Learning (ML) model based on the learned representation.
An Efficient and Flexible Deep Learning Method for Signal Delineation via Keypoints Estimation
Jul 24, 2024



Abstract:Deep Learning (DL) methods have been used for electrocardiogram (ECG) processing in a wide variety of tasks, demonstrating good performance compared with traditional signal processing algorithms. These methods offer an efficient framework with a limited need for apriori data pre-processing and feature engineering. While several studies use this approach for ECG signal delineation, a significant gap persists between the expected and the actual outcome. Existing methods rely on a sample-to-sample classifier. However, the clinical expected outcome consists of a set of onset, offset, and peak for the different waves that compose each R-R interval. To align the actual with the expected output, it is necessary to incorporate post-processing algorithms. This counteracts two of the main advantages of DL models, since these algorithms are based on assumptions and slow down the method's performance. In this paper, we present Keypoint Estimation for Electrocardiogram Delineation (KEED), a novel DL model designed for keypoint estimation, which organically offers an output aligned with clinical expectations. By standing apart from the conventional sample-to-sample classifier, we achieve two benefits: (i) Eliminate the need for additional post-processing, and (ii) Establish a flexible framework that allows the adjustment of the threshold value considering the sensitivity-specificity tradeoff regarding the particular clinical requirements. The proposed method's performance is compared with state-of-the-art (SOTA) signal processing methods. Remarkably, KEED significantly outperforms despite being optimized with an extremely limited annotated data. In addition, KEED decreases the inference time by a factor ranging from 52x to 703x.
Learning Beyond Similarities: Incorporating Dissimilarities between Positive Pairs in Self-Supervised Time Series Learning
Sep 14, 2023



Abstract:By identifying similarities between successive inputs, Self-Supervised Learning (SSL) methods for time series analysis have demonstrated their effectiveness in encoding the inherent static characteristics of temporal data. However, an exclusive emphasis on similarities might result in representations that overlook the dynamic attributes critical for modeling cardiovascular diseases within a confined subject cohort. Introducing Distilled Encoding Beyond Similarities (DEBS), this paper pioneers an SSL approach that transcends mere similarities by integrating dissimilarities among positive pairs. The framework is applied to electrocardiogram (ECG) signals, leading to a notable enhancement of +10\% in the detection accuracy of Atrial Fibrillation (AFib) across diverse subjects. DEBS underscores the potential of attaining a more refined representation by encoding the dynamic characteristics of time series data, tapping into dissimilarities during the optimization process. Broadly, the strategy delineated in this study holds the promise of unearthing novel avenues for advancing SSL methodologies tailored to temporal data.
Subject-based Non-contrastive Self-Supervised Learning for ECG Signal Processing
May 12, 2023



Abstract:Extracting information from the electrocardiography (ECG) signal is an essential step in the design of digital health technologies in cardiology. In recent years, several machine learning (ML) algorithms for automatic extraction of information in ECG have been proposed. Supervised learning methods have successfully been used to identify specific aspects in the signal, like detection of rhythm disorders (arrhythmias). Self-supervised learning (SSL) methods, on the other hand, can be used to extract all the features contained in the data. The model is optimized without any specific goal and learns from the data itself. By adapting state-of-the-art computer vision methodologies to the signal processing domain, a few SSL approaches have been reported recently for ECG processing. However, such SSL methods require either data augmentation or negative pairs, which limits the method to only look for similarities between two ECG inputs, either two versions of the same signal or two signals from the same subject. This leads to models that are very effective at extracting characteristics that are stable in a subject, such as gender or age. But they are not successful at capturing changes within the ECG recording that can explain dynamic aspects, like different arrhythmias or different sleep stages. In this work, we introduce the first SSL method that uses neither data augmentation nor negative pairs for understanding ECG signals, and still, achieves comparable quality representations. As a result, it is possible to design a SSL method that not only captures similarities between two inputs, but also captures dissimilarities for a complete understanding of the data. In addition, a model based on transformer blocks is presented, which produces better results than a model based on convolutional layers (XResNet50) with almost the same number of parameters.
DENS-ECG: A Deep Learning Approach for ECG Signal Delineation
May 18, 2020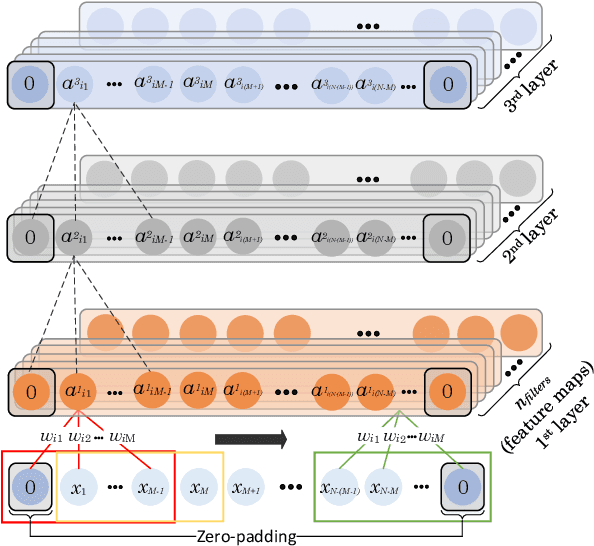

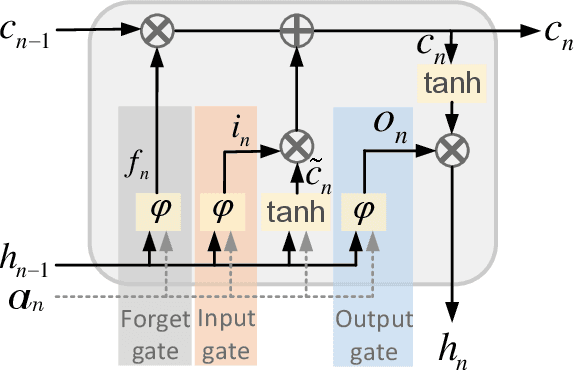
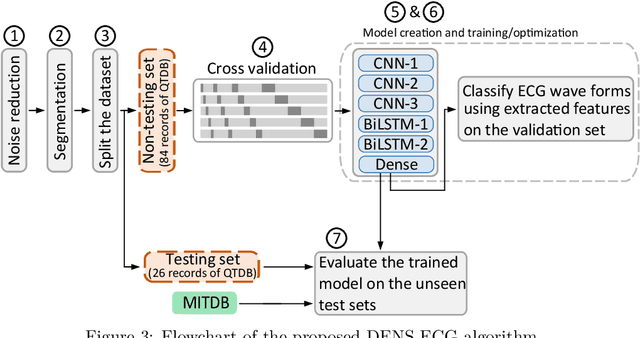
Abstract:Objectives: With the technological advancements in the field of tele-health monitoring, it is now possible to gather huge amounts of electro-physiological signals such as electrocardiogram (ECG). It is therefore necessary to develop models/algorithms that are capable of analysing these massive amounts of data in real-time. This paper proposes a deep learning model for real-time segmentation of heartbeats. Methods: The proposed algorithm, named as the DENS-ECG algorithm, combines convolutional neural network (CNN) and long short-term memory (LSTM) model to detect onset, peak, and offset of different heartbeat waveforms such as the P-wave, QRS complex, T-wave, and No wave (NW). Using ECG as the inputs, the model learns to extract high level features through the training process, which, unlike other classical machine learning based methods, eliminates the feature engineering step. Results: The proposed DENS-ECG model was trained and validated on a dataset with 105 ECGs of length 15 minutes each and achieved an average sensitivity and precision of 97.95% and 95.68%, respectively, using a 5-fold cross validation. Additionally, the model was evaluated on an unseen dataset to examine its robustness in QRS detection, which resulted in a sensitivity of 99.61% and precision of 99.52%. Conclusion: The empirical results show the flexibility and accuracy of the combined CNN-LSTM model for ECG signal delineation. Significance: This paper proposes an efficient and easy to use approach using deep learning for heartbeat segmentation, which could potentially be used in real-time tele-health monitoring systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge