Robin Sandkühler
Department of Biomedical Engineering, University of Basel, Basel, Switzerland
DeScarGAN: Disease-Specific Anomaly Detection with Weak Supervision
Jul 28, 2020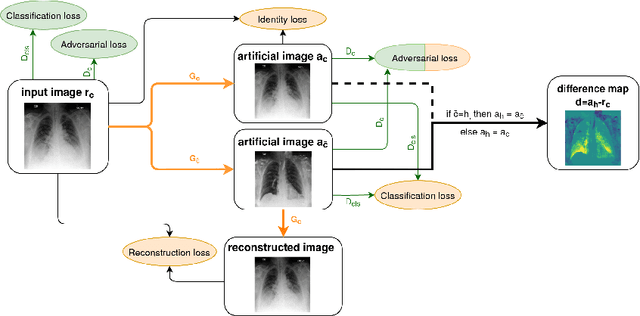
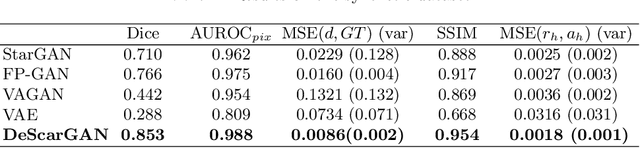
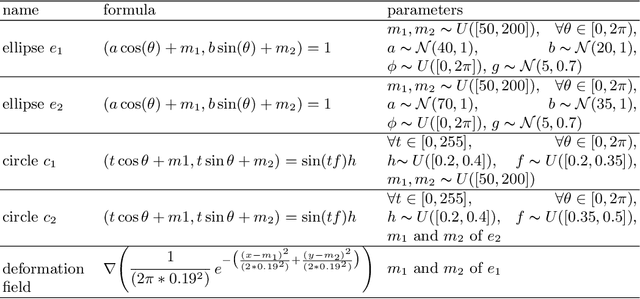
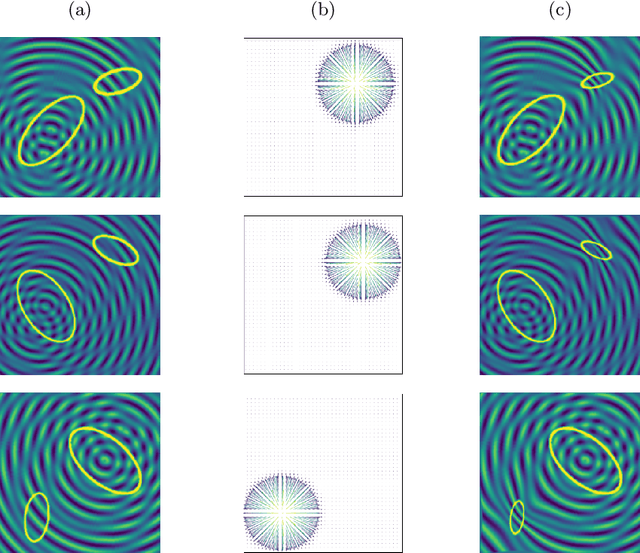
Abstract:Anomaly detection and localization in medical images is a challenging task, especially when the anomaly exhibits a change of existing structures, e.g., brain atrophy or changes in the pleural space due to pleural effusions. In this work, we present a weakly supervised and detail-preserving method that is able to detect structural changes of existing anatomical structures. In contrast to standard anomaly detection methods, our method extracts information about the disease characteristics from two groups: a group of patients affected by the same disease and a healthy control group. Together with identity-preserving mechanisms, this enables our method to extract highly disease-specific characteristics for a more detailed detection of structural changes. We designed a specific synthetic data set to evaluate and compare our method against state-of-the-art anomaly detection methods. Finally, we show the performance of our method on chest X-ray images. Our method called DeScarGAN outperforms other anomaly detection methods on the synthetic data set and by visual inspection on the chest X-ray image data set.
Accelerated Motion-Aware MR Imaging via Motion Prediction from K-Space Center
Aug 26, 2019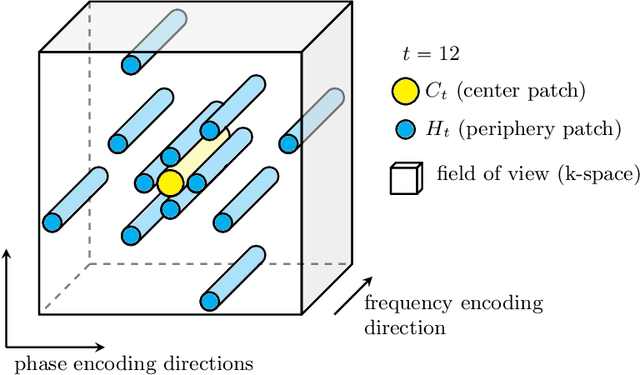
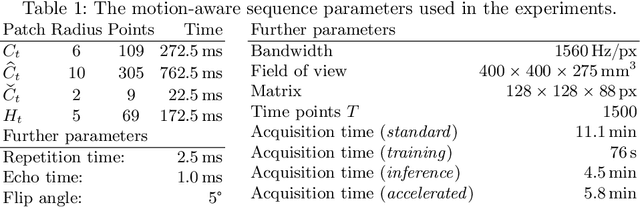
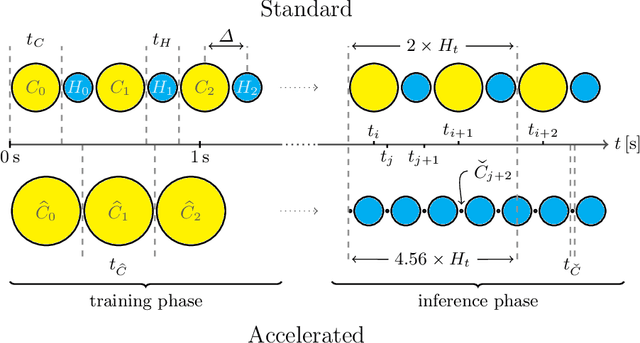
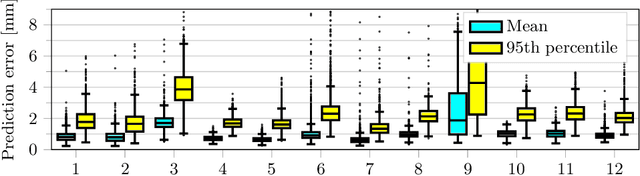
Abstract:Motion has been a challenge for magnetic resonance (MR) imaging ever since the MR has been invented. Especially in volumetric imaging of thoracic and abdominal organs, motion-awareness is essential for reducing motion artifacts in the final image. A recently proposed MR imaging approach copes with motion by observing the motion patterns during the acquisition. Repetitive scanning of the k-space center region enables the extraction of the patient motion while acquiring the remaining part of the k-space. Due to highly redundant measurements of the center, the required scanning time of over 11 min and the reconstruction time of 2 h exceed clinical applicability though. We propose an accelerated motion-aware MR imaging method where the motion is inferred from small-sized k-space center patches and an initial training phase during which the characteristic movements are modeled. Thereby, acquisition times are reduced by a factor of almost 2 and reconstruction times by two orders of magnitude. Moreover, we improve the existing motion-aware approach with a systematic temporal shift correction to achieve a sharper image reconstruction. We tested our method on 12 volunteers and scanned their lungs and abdomen under free breathing. We achieved equivalent to higher reconstruction quality using the motion-prediction compared to the slower existing approach.
Recurrent Registration Neural Networks for Deformable Image Registration
Jun 07, 2019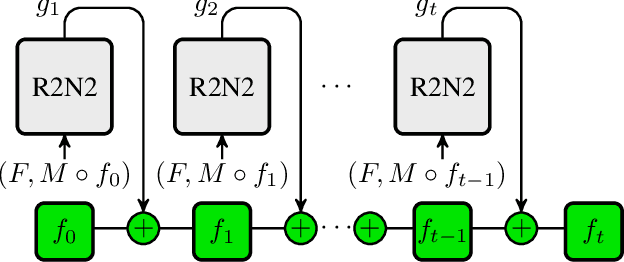
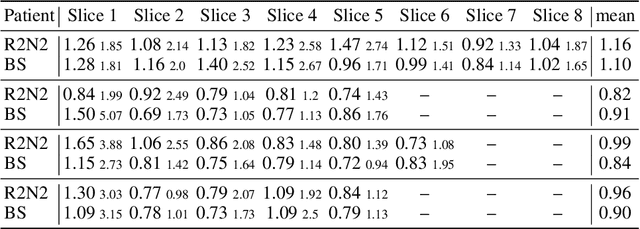
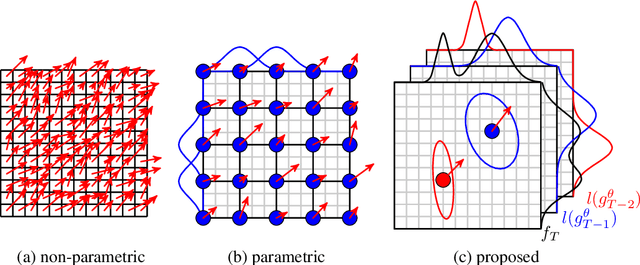
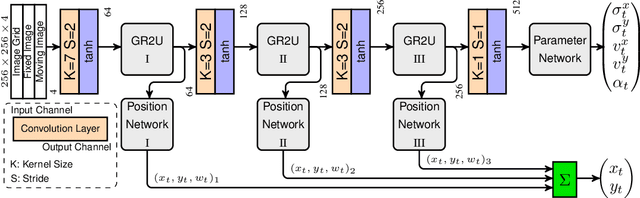
Abstract:Parametric spatial transformation models have been successfully applied to image registration tasks. In such models, the transformation of interest is parameterized by a fixed set of basis functions as for example B-splines. Each basis function is located on a fixed regular grid position among the image domain, because the transformation of interest is not known in advance. As a consequence, not all basis functions will necessarily contribute to the final transformation which results in a non-compact representation of the transformation. We reformulate the pairwise registration problem as a recursive sequence of successive alignments. For each element in the sequence, a local deformation defined by its position, shape, and weight is computed by our recurrent registration neural network. The sum of all local deformations yield the final spatial alignment of both images. Formulating the registration problem in this way allows the network to detect non-aligned regions in the images and to learn how to locally refine the registration properly. In contrast to current non-sequence-based registration methods, our approach iteratively applies local spatial deformations to the images until the desired registration accuracy is achieved. We trained our network on 2D magnetic resonance images of the lung and compared our method to a standard parametric B-spline registration. The experiments show, that our method performs on par for the accuracy but yields a more compact representation of the transformation. Furthermore, we achieve a speedup of around 15 compared to the B-spline registration.
AirLab: Autograd Image Registration Laboratory
Jun 26, 2018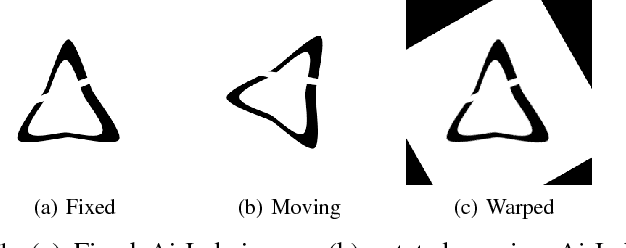
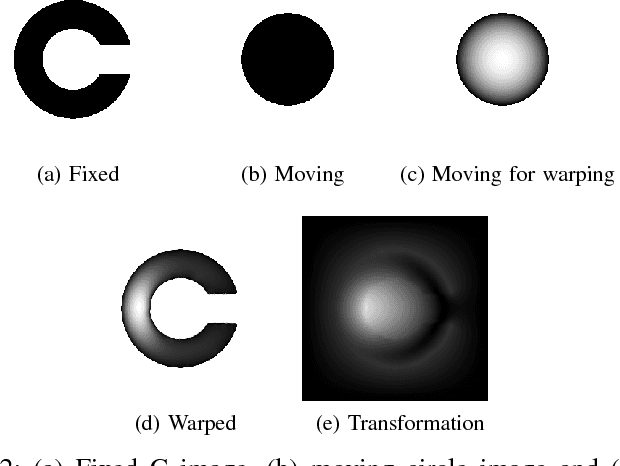
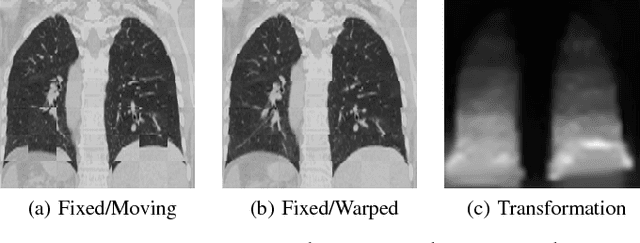
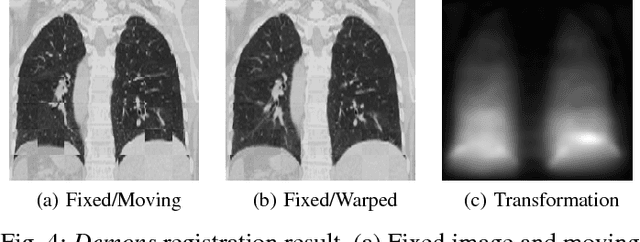
Abstract:Medical image registration is an active research topic and forms a basis for many medical image analysis tasks. Although image registration is a rather general concept specialized methods are usually required to target a specific registration problem. The development and implementation of such methods has been tough so far as the gradient of the objective has to be computed. Also, its evaluation has to be performed preferably on a GPU for larger images and for more complex transformation models and regularization terms. This hinders researchers from rapid prototyping and poses hurdles to reproduce research results. There is a clear need for an environment which hides this complexity to put the modeling and the experimental exploration of registration methods into the foreground. With the "Autograd Image Registration Laboratory" (AirLab), we introduce an open laboratory for image registration tasks, where the analytic gradients of the objective function are computed automatically and the device where the computations are performed, on a CPU or a GPU, is transparent. It is meant as a laboratory for researchers and developers enabling them to rapidly try out new ideas for registering images and to reproduce registration results which have already been published. AirLab is implemented in Python using PyTorch as tensor and optimization library and SimpleITK for basic image IO. Therefore, it profits from recent advances made by the machine learning community concerning optimization and deep neural network models. The present draft of this paper roughly outlines AirLab with first code snippets and performance analyses. A more exhaustive introduction will follow as a final version soon.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge