Parsa Esfahanian
Coronary Artery Segmentation and Vessel-Type Classification in X-Ray Angiography
Jan 24, 2026Abstract:X-ray coronary angiography (XCA) is the clinical reference standard for assessing coronary artery disease, yet quantitative analysis is limited by the difficulty of robust vessel segmentation in routine data. Low contrast, motion, foreshortening, overlap, and catheter confounding degrade segmentation and contribute to domain shift across centers. Reliable segmentation, together with vessel-type labeling, enables vessel-specific coronary analytics and downstream measurements that depend on anatomical localization. From 670 cine sequences (407 subjects), we select a best frame near peak opacification using a low-intensity histogram criterion and apply joint super-resolution and enhancement. We benchmark classical Meijering, Frangi, and Sato vesselness filters under per-image oracle tuning, a single global mean setting, and per-image parameter prediction via Support Vector Regression (SVR). Neural baselines include U-Net, FPN, and a Swin Transformer, trained with coronary-only and merged coronary+catheter supervision. A second stage assigns vessel identity (LAD, LCX, RCA). External evaluation uses the public DCA1 cohort. SVR per-image tuning improves Dice over global means for all classical filters (e.g., Frangi: 0.759 vs. 0.741). Among deep models, FPN attains 0.914+/-0.007 Dice (coronary-only), and merged coronary+catheter labels further improve to 0.931+/-0.006. On DCA1 as a strict external test, Dice drops to 0.798 (coronary-only) and 0.814 (merged), while light in-domain fine-tuning recovers to 0.881+/-0.014 and 0.882+/-0.015. Vessel-type labeling achieves 98.5% accuracy (Dice 0.844) for RCA, 95.4% (0.786) for LAD, and 96.2% (0.794) for LCX. Learned per-image tuning strengthens classical pipelines, while high-resolution FPN models and merged-label supervision improve stability and external transfer with modest adaptation.
GACNN: Training Deep Convolutional Neural Networks with Genetic Algorithm
Sep 29, 2019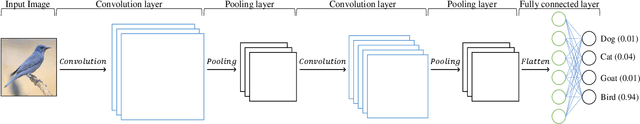
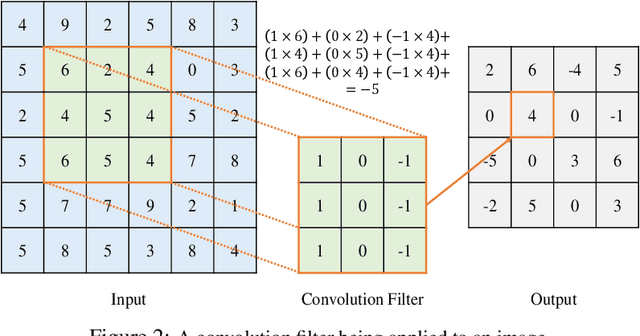
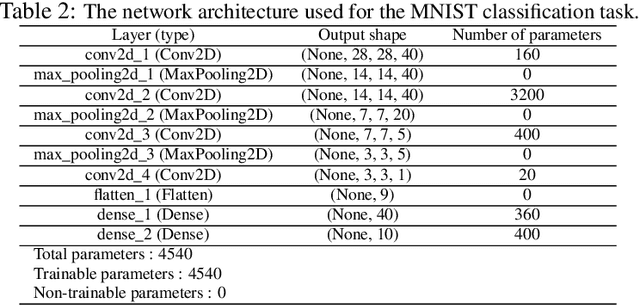
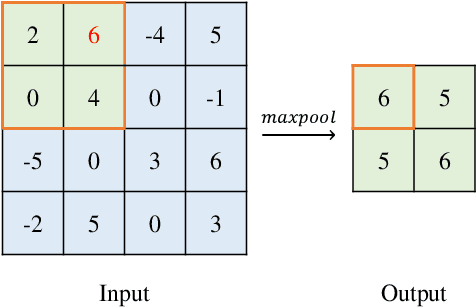
Abstract:Convolutional Neural Networks (CNNs) have gained a significant attraction in the recent years due to their increasing real-world applications. Their performance is highly dependent to the network structure and the selected optimization method for tuning the network parameters. In this paper, we propose novel yet efficient methods for training convolutional neural networks. The most of current state of the art learning method for CNNs are based on Gradient decent. In contrary to the traditional CNN training methods, we propose to optimize the CNNs using methods based on Genetic Algorithms (GAs). These methods are carried out using three individual GA schemes, Steady-State, Generational, and Elitism. We present new genetic operators for crossover, mutation and also an innovative encoding paradigm of CNNs to chromosomes aiming to reduce the resulting chromosome's size by a large factor. We compare the effectiveness and scalability of our encoding with the traditional encoding. Furthermore, the performance of individual GA schemes used for training the networks were compared with each other in means of convergence rate and overall accuracy. Finally, our new encoding alongside the superior GA-based training scheme is compared to Backpropagation training with Adam optimization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge