Omer Bayraktar
3D-Guided Scalable Flow Matching for Generating Volumetric Tissue Spatial Transcriptomics from Serial Histology
Nov 18, 2025Abstract:A scalable and robust 3D tissue transcriptomics profile can enable a holistic understanding of tissue organization and provide deeper insights into human biology and disease. Most predictive algorithms that infer ST directly from histology treat each section independently and ignore 3D structure, while existing 3D-aware approaches are not generative and do not scale well. We present Holographic Tissue Expression Inpainting and Analysis (HoloTea), a 3D-aware flow-matching framework that imputes spot-level gene expression from H&E while explicitly using information from adjacent sections. Our key idea is to retrieve morphologically corresponding spots on neighboring slides in a shared feature space and fuse this cross section context into a lightweight ControlNet, allowing conditioning to follow anatomical continuity. To better capture the count nature of the data, we introduce a 3D-consistent prior for flow matching that combines a learned zero-inflated negative binomial (ZINB) prior with a spatial-empirical prior constructed from neighboring sections. A global attention block introduces 3D H&E scaling linearly with the number of spots in the slide, enabling training and inference on large 3D ST datasets. Across three spatial transcriptomics datasets spanning different tissue types and resolutions, HoloTea consistently improves 3D expression accuracy and generalization compared to 2D and 3D baselines. We envision HoloTea advancing the creation of accurate 3D virtual tissues, ultimately accelerating biomarker discovery and deepening our understanding of disease.
Noise2Stack: Improving Image Restoration by Learning from Volumetric Data
Nov 10, 2020
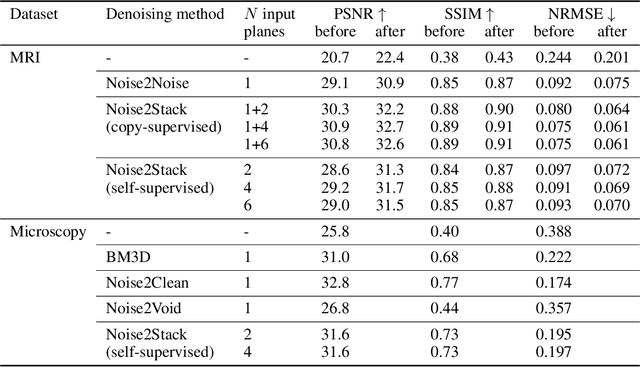
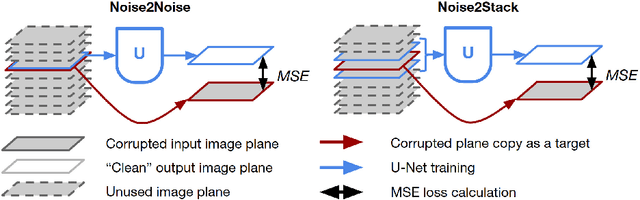
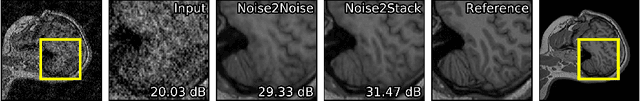
Abstract:Biomedical images are noisy. The imaging equipment itself has physical limitations, and the consequent experimental trade-offs between signal-to-noise ratio, acquisition speed, and imaging depth exacerbate the problem. Denoising is, therefore, an essential part of any image processing pipeline, and convolutional neural networks are currently the method of choice for this task. One popular approach, Noise2Noise, does not require clean ground truth, and instead, uses a second noisy copy as a training target. Self-supervised methods, like Noise2Self and Noise2Void, relax data requirements by learning the signal without an explicit target but are limited by the lack of information in a single image. Here, we introduce Noise2Stack, an extension of the Noise2Noise method to image stacks that takes advantage of a shared signal between spatially neighboring planes. Our experiments on magnetic resonance brain scans and newly acquired multiplane microscopy data show that learning only from image neighbors in a stack is sufficient to outperform Noise2Noise and Noise2Void and close the gap to supervised denoising methods. Our findings point towards low-cost, high-reward improvement in the denoising pipeline of multiplane biomedical images. As a part of this work, we release a microscopy dataset to establish a benchmark for the multiplane image denoising.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge