Nicolas Pinon
MYRIAD
GAN-based synthetic FDG PET images from T1 brain MRI can serve to improve performance of deep unsupervised anomaly detection models
May 12, 2025Abstract:Background and Objective. Research in the cross-modal medical image translation domain has been very productive over the past few years in tackling the scarce availability of large curated multimodality datasets with the promising performance of GAN-based architectures. However, only a few of these studies assessed task-based related performance of these synthetic data, especially for the training of deep models. Method. We design and compare different GAN-based frameworks for generating synthetic brain [18F]fluorodeoxyglucose (FDG) PET images from T1 weighted MRI data. We first perform standard qualitative and quantitative visual quality evaluation. Then, we explore further impact of using these fake PET data in the training of a deep unsupervised anomaly detection (UAD) model designed to detect subtle epilepsy lesions in T1 MRI and FDG PET images. We introduce novel diagnostic task-oriented quality metrics of the synthetic FDG PET data tailored to our unsupervised detection task, then use these fake data to train a use case UAD model combining a deep representation learning based on siamese autoencoders with a OC-SVM density support estimation model. This model is trained on normal subjects only and allows the detection of any variation from the pattern of the normal population. We compare the detection performance of models trained on 35 paired real MR T1 of normal subjects paired either on 35 true PET images or on 35 synthetic PET images generated from the best performing generative models. Performance analysis is conducted on 17 exams of epilepsy patients undergoing surgery. Results. The best performing GAN-based models allow generating realistic fake PET images of control subject with SSIM and PSNR values around 0.9 and 23.8, respectively and in distribution (ID) with regard to the true control dataset. The best UAD model trained on these synthetic normative PET data allows reaching 74% sensitivity. Conclusion. Our results confirm that GAN-based models are the best suited for MR T1 to FDG PET translation, outperforming transformer or diffusion models. We also demonstrate the diagnostic value of these synthetic data for the training of UAD models and evaluation on clinical exams of epilepsy patients. Our code and the normative image dataset are available.
Anomaly detection in image or latent space of patch-based auto-encoders for industrial image analysis
Jul 04, 2023Abstract:We study several methods for detecting anomalies in color images, constructed on patch-based auto-encoders. Wecompare the performance of three types of methods based, first, on the error between the original image and its reconstruction,second, on the support estimation of the normal image distribution in the latent space, and third, on the error between the originalimage and a restored version of the reconstructed image. These methods are evaluated on the industrial image database MVTecADand compared to two competitive state-of-the-art methods.
One-Class SVM on siamese neural network latent space for Unsupervised Anomaly Detection on brain MRI White Matter Hyperintensities
Apr 17, 2023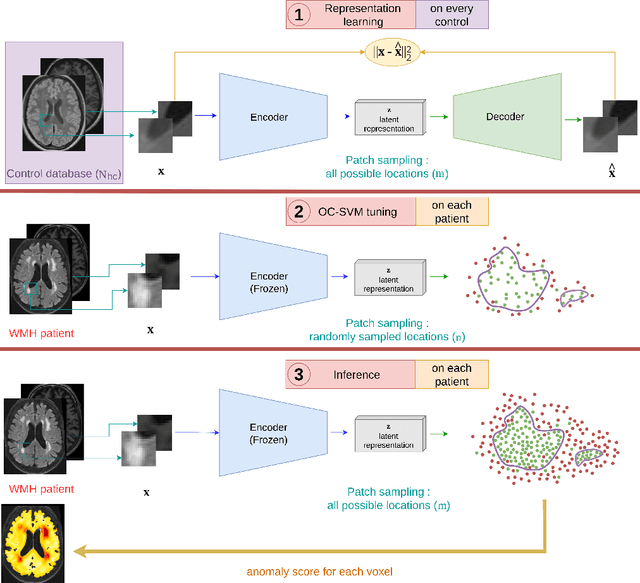
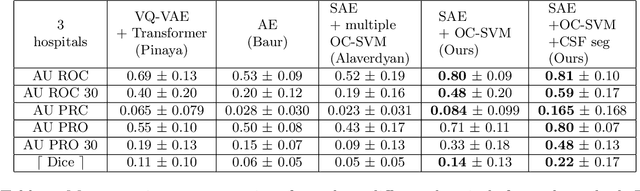
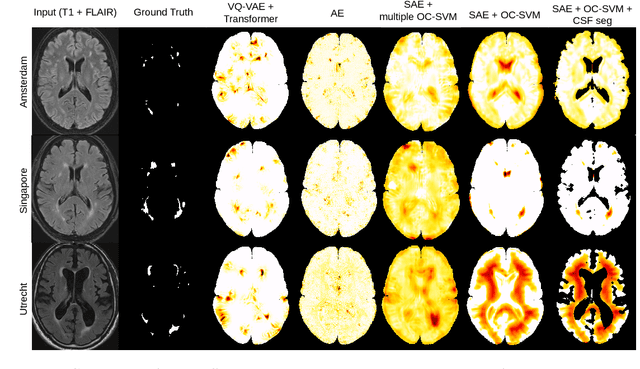
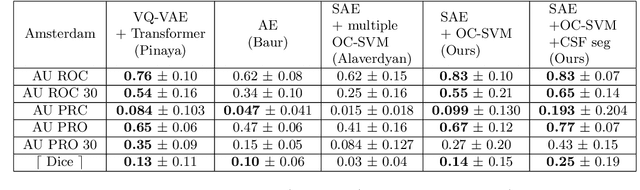
Abstract:Anomaly detection remains a challenging task in neuroimaging when little to no supervision is available and when lesions can be very small or with subtle contrast. Patch-based representation learning has shown powerful representation capacities when applied to industrial or medical imaging and outlier detection methods have been applied successfully to these images. In this work, we propose an unsupervised anomaly detection (UAD) method based on a latent space constructed by a siamese patch-based auto-encoder and perform the outlier detection with a One-Class SVM training paradigm tailored to the lesion detection task in multi-modality neuroimaging. We evaluate performances of this model on a public database, the White Matter Hyperintensities (WMH) challenge and show in par performance with the two best performing state-of-the-art methods reported so far.
Brain subtle anomaly detection based on auto-encoders latent space analysis : application to de novo parkinson patients
Feb 27, 2023

Abstract:Neural network-based anomaly detection remains challenging in clinical applications with little or no supervised information and subtle anomalies such as hardly visible brain lesions. Among unsupervised methods, patch-based auto-encoders with their efficient representation power provided by their latent space, have shown good results for visible lesion detection. However, the commonly used reconstruction error criterion may limit their performance when facing less obvious lesions. In this work, we design two alternative detection criteria. They are derived from multivariate analysis and can more directly capture information from latent space representations. Their performance compares favorably with two additional supervised learning methods, on a difficult de novo Parkinson Disease (PD) classification task.
Patch vs. Global Image-Based Unsupervised Anomaly Detection in MR Brain Scans of Early Parkinsonian Patients
Oct 25, 2021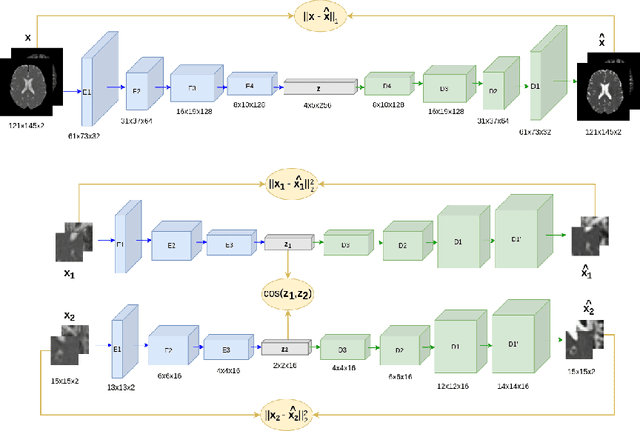
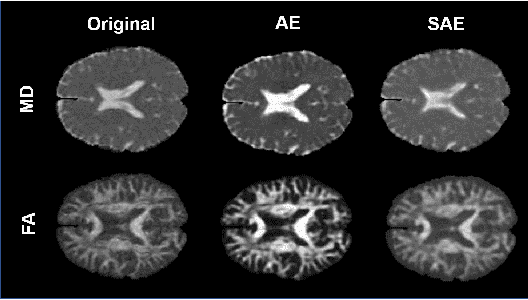
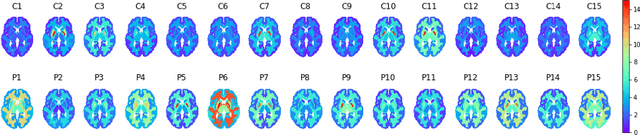
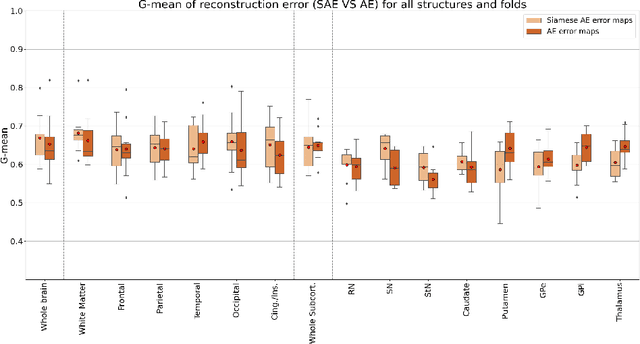
Abstract:Although neural networks have proven very successful in a number of medical image analysis applications, their use remains difficult when targeting subtle tasks such as the identification of barely visible brain lesions, especially given the lack of annotated datasets. Good candidate approaches are patch-based unsupervised pipelines which have both the advantage to increase the number of input data and to capture local and fine anomaly patterns distributed in the image, while potential inconveniences are the loss of global structural information. We illustrate this trade-off on Parkinson's disease (PD) anomaly detection comparing the performance of two anomaly detection models based on a spatial auto-encoder (AE) and an adaptation of a patch-fed siamese auto-encoder (SAE). On average, the SAE model performs better, showing that patches may indeed be advantageous.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge