Naveen Sivadasan
Multi-label classification for biomedical literature: an overview of the BioCreative VII LitCovid Track for COVID-19 literature topic annotations
Apr 20, 2022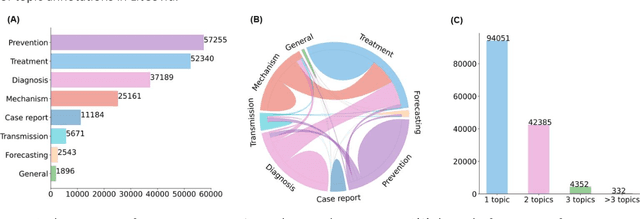
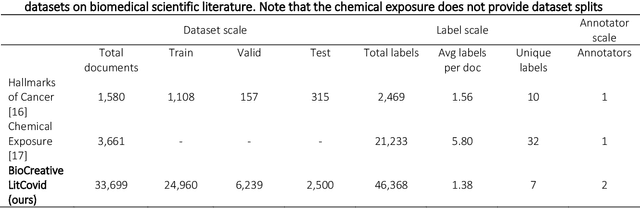
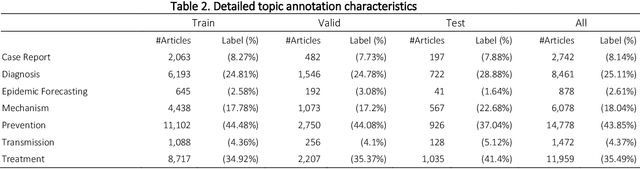
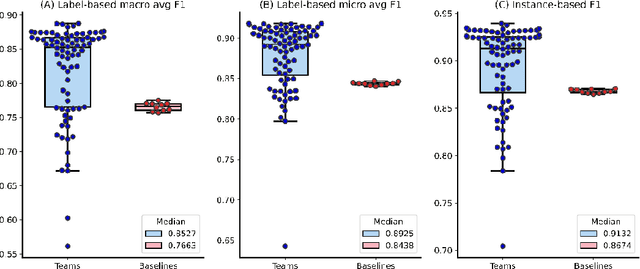
Abstract:The COVID-19 pandemic has been severely impacting global society since December 2019. Massive research has been undertaken to understand the characteristics of the virus and design vaccines and drugs. The related findings have been reported in biomedical literature at a rate of about 10,000 articles on COVID-19 per month. Such rapid growth significantly challenges manual curation and interpretation. For instance, LitCovid is a literature database of COVID-19-related articles in PubMed, which has accumulated more than 200,000 articles with millions of accesses each month by users worldwide. One primary curation task is to assign up to eight topics (e.g., Diagnosis and Treatment) to the articles in LitCovid. Despite the continuing advances in biomedical text mining methods, few have been dedicated to topic annotations in COVID-19 literature. To close the gap, we organized the BioCreative LitCovid track to call for a community effort to tackle automated topic annotation for COVID-19 literature. The BioCreative LitCovid dataset, consisting of over 30,000 articles with manually reviewed topics, was created for training and testing. It is one of the largest multilabel classification datasets in biomedical scientific literature. 19 teams worldwide participated and made 80 submissions in total. Most teams used hybrid systems based on transformers. The highest performing submissions achieved 0.8875, 0.9181, and 0.9394 for macro F1-score, micro F1-score, and instance-based F1-score, respectively. The level of participation and results demonstrate a successful track and help close the gap between dataset curation and method development. The dataset is publicly available via https://ftp.ncbi.nlm.nih.gov/pub/lu/LitCovid/biocreative/ for benchmarking and further development.
Adiabatic Quantum Feature Selection for Sparse Linear Regression
Jun 04, 2021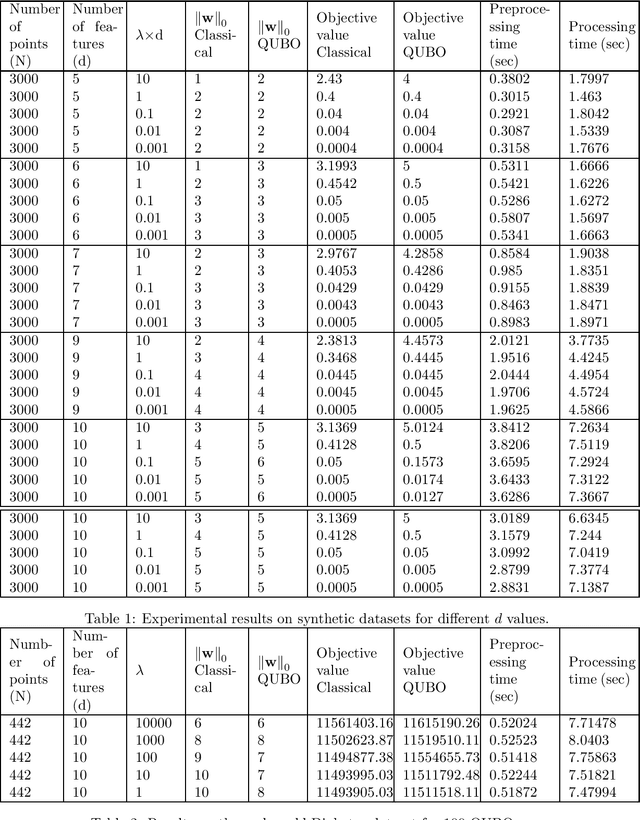
Abstract:Linear regression is a popular machine learning approach to learn and predict real valued outputs or dependent variables from independent variables or features. In many real world problems, its beneficial to perform sparse linear regression to identify important features helpful in predicting the dependent variable. It not only helps in getting interpretable results but also avoids overfitting when the number of features is large, and the amount of data is small. The most natural way to achieve this is by using `best subset selection' which penalizes non-zero model parameters by adding $\ell_0$ norm over parameters to the least squares loss. However, this makes the objective function non-convex and intractable even for a small number of features. This paper aims to address the intractability of sparse linear regression with $\ell_0$ norm using adiabatic quantum computing, a quantum computing paradigm that is particularly useful for solving optimization problems faster. We formulate the $\ell_0$ optimization problem as a Quadratic Unconstrained Binary Optimization (QUBO) problem and solve it using the D-Wave adiabatic quantum computer. We study and compare the quality of QUBO solution on synthetic and real world datasets. The results demonstrate the effectiveness of the proposed adiabatic quantum computing approach in finding the optimal solution. The QUBO solution matches the optimal solution for a wide range of sparsity penalty values across the datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge