Mohamed Abdelmoniem
Panther: Faster and Cheaper Computations with Randomized Numerical Linear Algebra
Jan 21, 2026Abstract:Training modern deep learning models is increasingly constrained by GPU memory and compute limits. While Randomized Numerical Linear Algebra (RandNLA) offers proven techniques to compress these models, the lack of a unified, production-grade library prevents widely adopting these methods. We present Panther, a PyTorch-compatible library that consolidates established RandNLA algorithms into a single high-performance framework. Panther engineers efficient, drop-in replacements for standard components including sketched linear layers, 2D convolution, multi-head attention, and randomized matrix decompositions (such as pivoted CholeskyQR). By implementing a custom C++/CUDA backend (pawX), Panther provides an optimized implementation that can run on both CPUs and GPUs. We demonstrate the effectiveness of RandNLA techniques and Panther's ease of adoption. By replacing standard PyTorch linear layers with Panther layers (requiring only a few lines of code) we achieve significant memory savings (up to 75%) on BERT while maintaining comparable loss. Source code is available (MIT License) at https://github.com/FahdSeddik/panther, along with demonstration video at https://youtu.be/7M3RQb4KWxs.
Evaluating Performance of Machine Learning Models for Diabetic Sensorimotor Polyneuropathy Severity Classification using Biomechanical Signals during Gait
May 21, 2022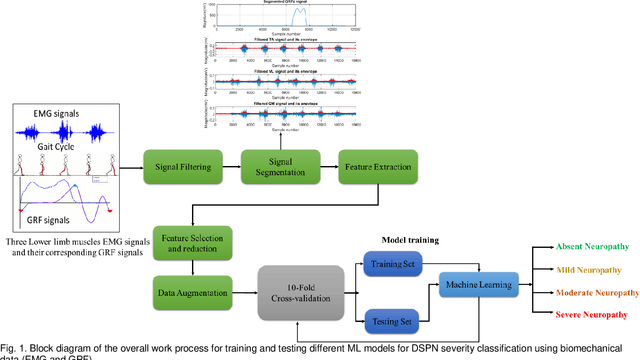
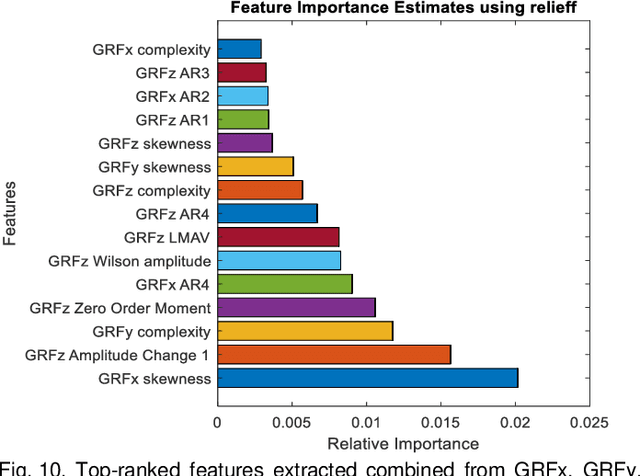
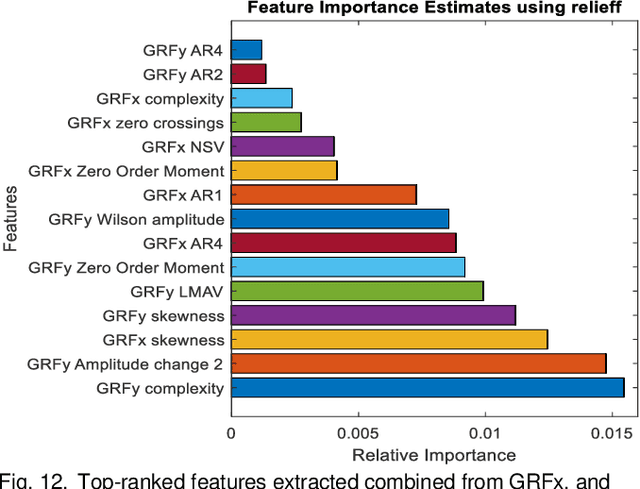
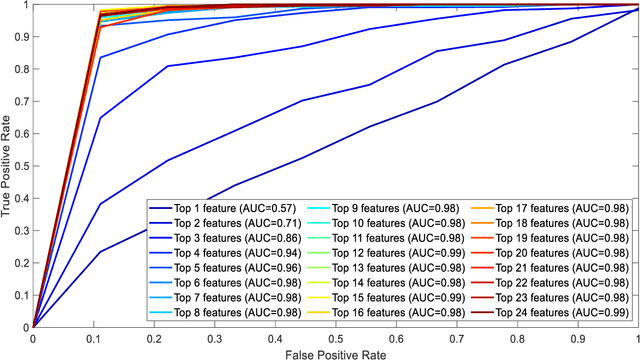
Abstract:Diabetic sensorimotor polyneuropathy (DSPN) is one of the prevalent forms of neuropathy affected by diabetic patients that involves alterations in biomechanical changes in human gait. In literature, for the last 50 years, researchers are trying to observe the biomechanical changes due to DSPN by studying muscle electromyography (EMG), and ground reaction forces (GRF). However, the literature is contradictory. In such a scenario, we are proposing to use Machine learning techniques to identify DSPN patients by using EMG, and GRF data. We have collected a dataset consists of three lower limb muscles EMG (tibialis anterior (TA), vastus lateralis (VL), gastrocnemius medialis (GM) and 3-dimensional GRF components (GRFx, GRFy, and GRFz). Raw EMG and GRF signals were preprocessed, and a newly proposed feature extraction technique scheme from literature was applied to extract the best features from the signals. The extracted feature list was ranked using Relief feature ranking techniques, and highly correlated features were removed. We have trained different ML models to find out the best-performing model and optimized that model. We trained the optimized ML models for different combinations of muscles and GRF components features, and the performance matrix was evaluated. This study has found ensemble classifier model was performing in identifying DSPN Severity, and we optimized it before training. For EMG analysis, we have found the best accuracy of 92.89% using the Top 14 features for features from GL, VL and TA muscles combined. In the GRF analysis, the model showed 94.78% accuracy by using the Top 15 features for the feature combinations extracted from GRFx, GRFy and GRFz signals. The performance of ML-based DSPN severity classification models, improved significantly, indicating their reliability in DSPN severity classification, for biomechanical data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge