Minh H. Vu
Using Synthetic Images to Augment Small Medical Image Datasets
Mar 02, 2025



Abstract:Recent years have witnessed a growing academic and industrial interest in deep learning (DL) for medical imaging. To perform well, DL models require very large labeled datasets. However, most medical imaging datasets are small, with a limited number of annotated samples. The reason they are small is usually because delineating medical images is time-consuming and demanding for oncologists. There are various techniques that can be used to augment a dataset, for example, to apply affine transformations or elastic transformations to available images, or to add synthetic images generated by a Generative Adversarial Network (GAN). In this work, we have developed a novel conditional variant of a current GAN method, the StyleGAN2, to generate multi-modal high-resolution medical images with the purpose to augment small medical imaging datasets with these synthetic images. We use the synthetic and real images from six datasets to train models for the downstream task of semantic segmentation. The quality of the generated medical images and the effect of this augmentation on the segmentation performance were evaluated afterward. Finally, the results indicate that the downstream segmentation models did not benefit from the generated images. Further work and analyses are required to establish how this augmentation affects the segmentation performance.
A Correlation- and Mean-Aware Loss Function and Benchmarking Framework to Improve GAN-based Tabular Data Synthesis
May 27, 2024Abstract:Advancements in science rely on data sharing. In medicine, where personal data are often involved, synthetic tabular data generated by generative adversarial networks (GANs) offer a promising avenue. However, existing GANs struggle to capture the complexities of real-world tabular data, which often contain a mix of continuous and categorical variables with potential imbalances and dependencies. We propose a novel correlation- and mean-aware loss function designed to address these challenges as a regularizer for GANs. To ensure a rigorous evaluation, we establish a comprehensive benchmarking framework using ten real-world datasets and eight established tabular GAN baselines. The proposed loss function demonstrates statistically significant improvements over existing methods in capturing the true data distribution, significantly enhancing the quality of synthetic data generated with GANs. The benchmarking framework shows that the enhanced synthetic data quality leads to improved performance in downstream machine learning (ML) tasks, ultimately paving the way for easier data sharing.
LatentAugment: Data Augmentation via Guided Manipulation of GAN's Latent Space
Jul 21, 2023



Abstract:Data Augmentation (DA) is a technique to increase the quantity and diversity of the training data, and by that alleviate overfitting and improve generalisation. However, standard DA produces synthetic data for augmentation with limited diversity. Generative Adversarial Networks (GANs) may unlock additional information in a dataset by generating synthetic samples having the appearance of real images. However, these models struggle to simultaneously address three key requirements: fidelity and high-quality samples; diversity and mode coverage; and fast sampling. Indeed, GANs generate high-quality samples rapidly, but have poor mode coverage, limiting their adoption in DA applications. We propose LatentAugment, a DA strategy that overcomes the low diversity of GANs, opening up for use in DA applications. Without external supervision, LatentAugment modifies latent vectors and moves them into latent space regions to maximise the synthetic images' diversity and fidelity. It is also agnostic to the dataset and the downstream task. A wide set of experiments shows that LatentAugment improves the generalisation of a deep model translating from MRI-to-CT beating both standard DA as well GAN-based sampling. Moreover, still in comparison with GAN-based sampling, LatentAugment synthetic samples show superior mode coverage and diversity. Code is available at: https://github.com/ltronchin/LatentAugment.
QU-BraTS: MICCAI BraTS 2020 Challenge on Quantifying Uncertainty in Brain Tumor Segmentation -- Analysis of Ranking Metrics and Benchmarking Results
Dec 19, 2021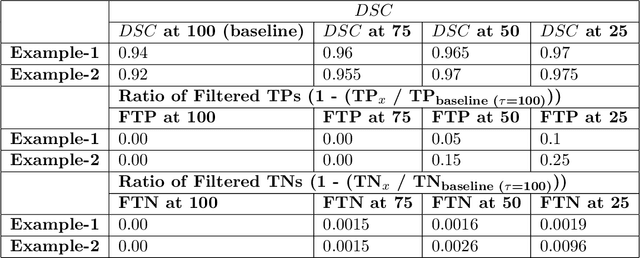
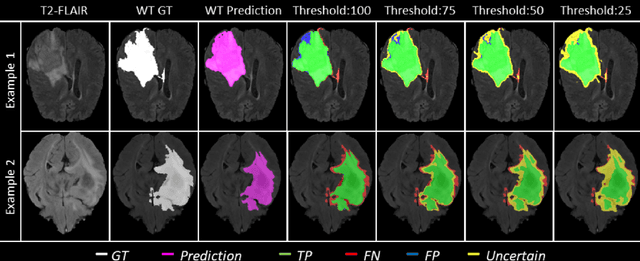
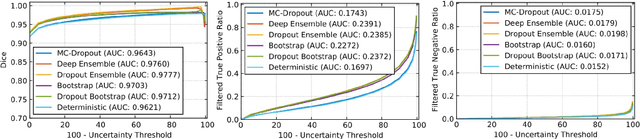
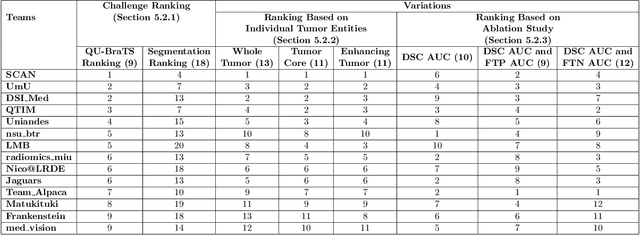
Abstract:Deep learning (DL) models have provided the state-of-the-art performance in a wide variety of medical imaging benchmarking challenges, including the Brain Tumor Segmentation (BraTS) challenges. However, the task of focal pathology multi-compartment segmentation (e.g., tumor and lesion sub-regions) is particularly challenging, and potential errors hinder the translation of DL models into clinical workflows. Quantifying the reliability of DL model predictions in the form of uncertainties, could enable clinical review of the most uncertain regions, thereby building trust and paving the way towards clinical translation. Recently, a number of uncertainty estimation methods have been introduced for DL medical image segmentation tasks. Developing metrics to evaluate and compare the performance of uncertainty measures will assist the end-user in making more informed decisions. In this study, we explore and evaluate a metric developed during the BraTS 2019-2020 task on uncertainty quantification (QU-BraTS), and designed to assess and rank uncertainty estimates for brain tumor multi-compartment segmentation. This metric (1) rewards uncertainty estimates that produce high confidence in correct assertions, and those that assign low confidence levels at incorrect assertions, and (2) penalizes uncertainty measures that lead to a higher percentages of under-confident correct assertions. We further benchmark the segmentation uncertainties generated by 14 independent participating teams of QU-BraTS 2020, all of which also participated in the main BraTS segmentation task. Overall, our findings confirm the importance and complementary value that uncertainty estimates provide to segmentation algorithms, and hence highlight the need for uncertainty quantification in medical image analyses. Our evaluation code is made publicly available at https://github.com/RagMeh11/QU-BraTS.
A Data-Adaptive Loss Function for Incomplete Data and Incremental Learning in Semantic Image Segmentation
Apr 22, 2021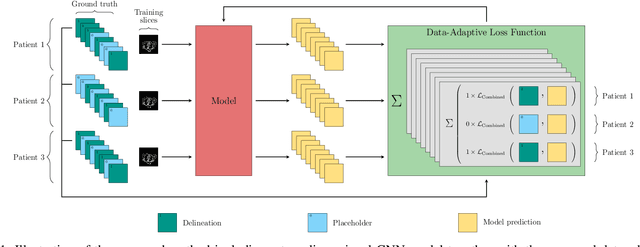
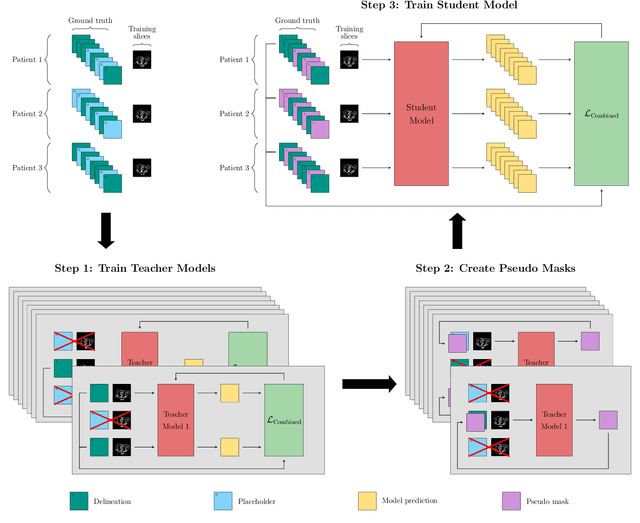
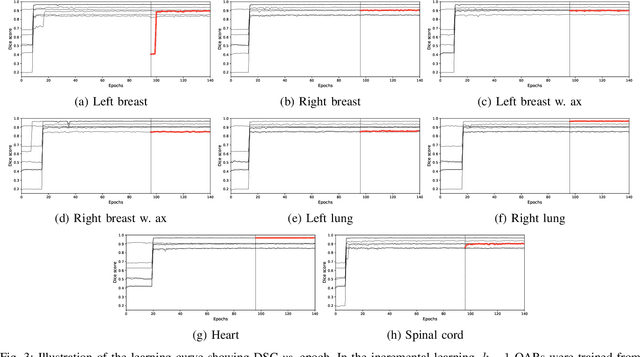
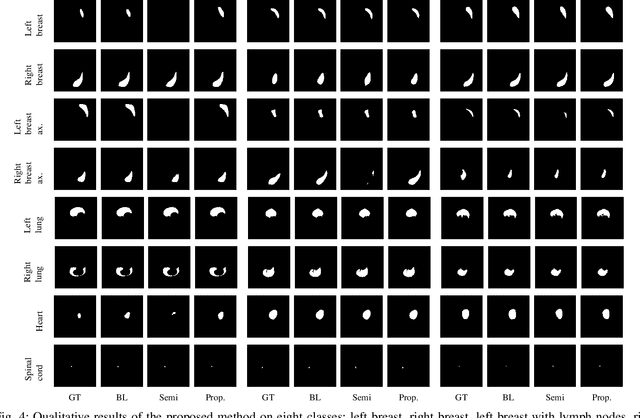
Abstract:In the last years, deep learning has dramatically improved the performances in a variety of medical image analysis applications. Among different types of deep learning models, convolutional neural networks have been among the most successful and they have been used in many applications in medical imaging. Training deep convolutional neural networks often requires large amounts of image data to generalize well to new unseen images. It is often time-consuming and expensive to collect large amounts of data in the medical image domain due to expensive imaging systems, and the need for experts to manually make ground truth annotations. A potential problem arises if new structures are added when a decision support system is already deployed and in use. Since the field of radiation therapy is constantly developing, the new structures would also have to be covered by the decision support system. In the present work, we propose a novel loss function, that adapts to the available data in order to utilize all available data, even when some have missing annotations. We demonstrate that the proposed loss function also works well in an incremental learning setting, where it can automatically incorporate new structures as they appear. Experiments on a large in-house data set show that the proposed method performs on par with baseline models, while greatly reducing the training time.
Multi-Decoder Networks with Multi-Denoising Inputs for Tumor Segmentation
Nov 16, 2020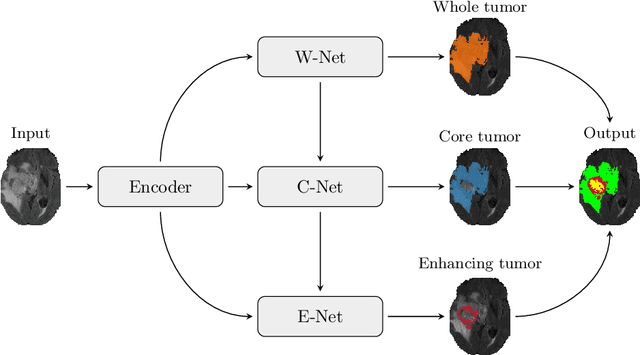
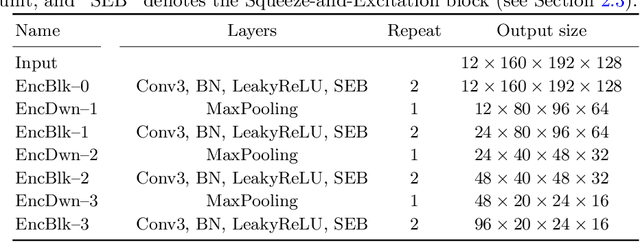
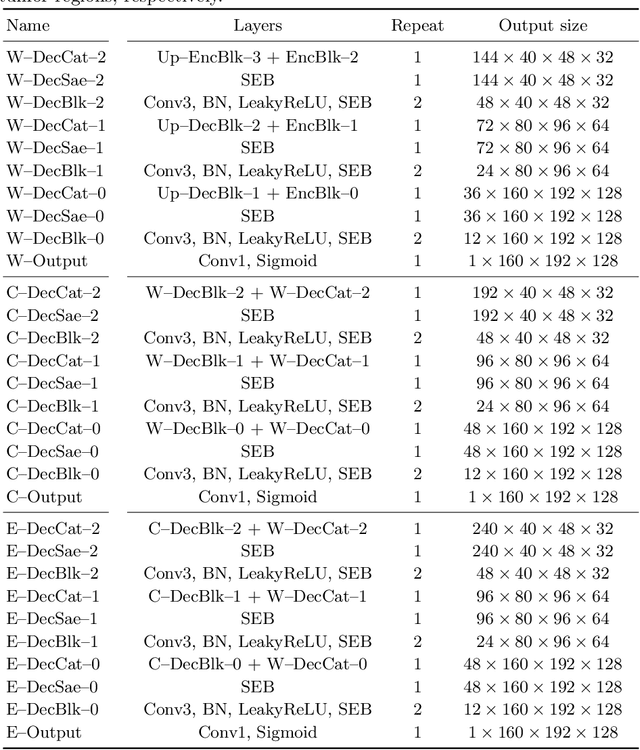
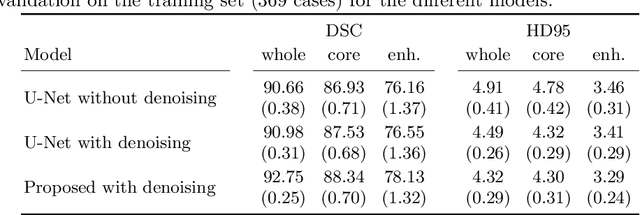
Abstract:Automatic segmentation of brain glioma from multimodal MRI scans plays a key role in clinical trials and practice. Unfortunately, manual segmentation is very challenging, time-consuming, costly, and often inaccurate despite human expertise due to the high variance and high uncertainty in the human annotations. In the present work, we develop an end-to-end deep-learning-based segmentation method using a multi-decoder architecture by jointly learning three separate sub-problems using a partly shared encoder. We also propose to apply smoothing methods to the input images to generate denoised versions as additional inputs to the network. The validation performance indicate an improvement when using the proposed method. The proposed method was ranked 2nd in the task of Quantification of Uncertainty in Segmentation in the Brain Tumors in Multimodal Magnetic Resonance Imaging Challenge 2020.
A Question-Centric Model for Visual Question Answering in Medical Imaging
Mar 02, 2020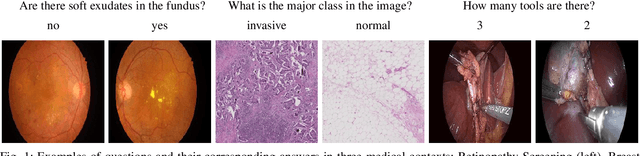
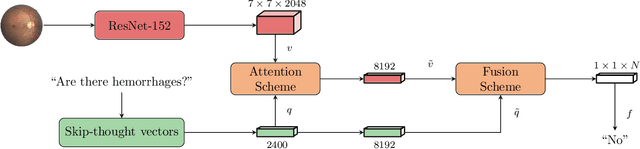
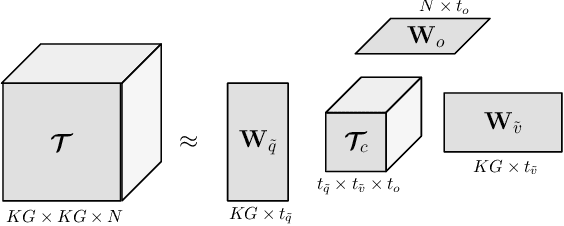
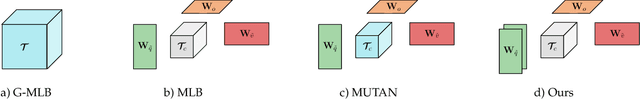
Abstract:Deep learning methods have proven extremely effective at performing a variety of medical image analysis tasks. With their potential use in clinical routine, their lack of transparency has however been one of their few weak points, raising concerns regarding their behavior and failure modes. While most research to infer model behavior has focused on indirect strategies that estimate prediction uncertainties and visualize model support in the input image space, the ability to explicitly query a prediction model regarding its image content offers a more direct way to determine the behavior of trained models. To this end, we present a novel Visual Question Answering approach that allows an image to be queried by means of a written question. Experiments on a variety of medical and natural image datasets show that by fusing image and question features in a novel way, the proposed approach achieves an equal or higher accuracy compared to current methods.
Evaluation of Multi-Slice Inputs to Convolutional Neural Networks for Medical Image Segmentation
Dec 22, 2019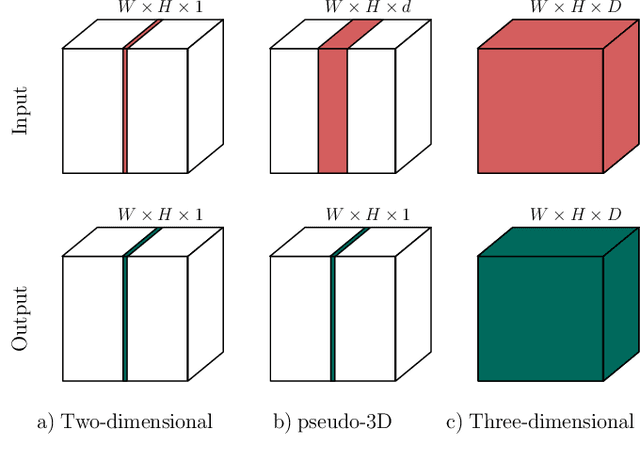
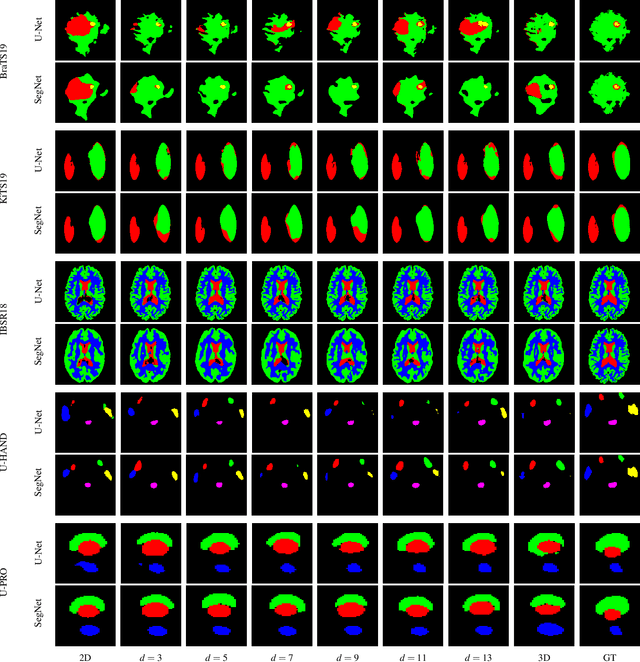
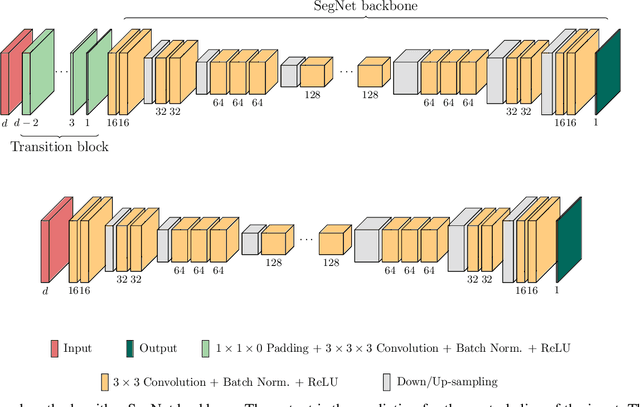
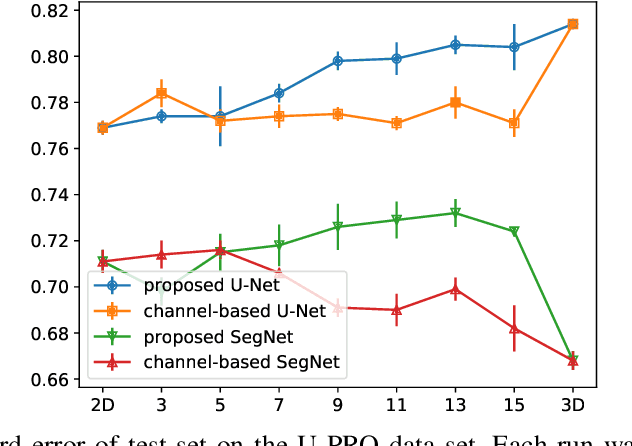
Abstract:When using Convolutional Neural Networks (CNNs) for segmentation of organs and lesions in medical images, the conventional approach is to work with inputs and outputs either as single slice (2D) or whole volumes (3D). One common alternative, in this study denoted as pseudo-3D, is to use a stack of adjacent slices as input and produce a prediction for at least the central slice. This approach gives the network the possibility to capture 3D spatial information, with only a minor additional computational cost. In this study, we systematically evaluate the segmentation performance and computational costs of this pseudo-3D approach as a function of the number of input slices, and compare the results to conventional end-to-end 2D and 3D CNNs. The standard pseudo-3D method regards the neighboring slices as multiple input image channels. We additionally evaluate a simple approach where the input stack is a volumetric input that is repeatably convolved in 3D to obtain a 2D feature map. This 2D map is in turn fed into a standard 2D network. We conducted experiments using two different CNN backbone architectures and on five diverse data sets covering different anatomical regions, imaging modalities, and segmentation tasks. We found that while both pseudo-3D methods can process a large number of slices at once and still be computationally much more efficient than fully 3D CNNs, a significant improvement over a regular 2D CNN was only observed for one of the five data sets. An analysis of the structural properties of the segmentation masks revealed no relations to the segmentation performance with respect to the number of input slices. The conclusion is therefore that in the general case, multi-slice inputs appear to not significantly improve segmentation results over using 2D or 3D CNNs.
End-to-End Cascaded U-Nets with a Localization Network for Kidney Tumor Segmentation
Oct 16, 2019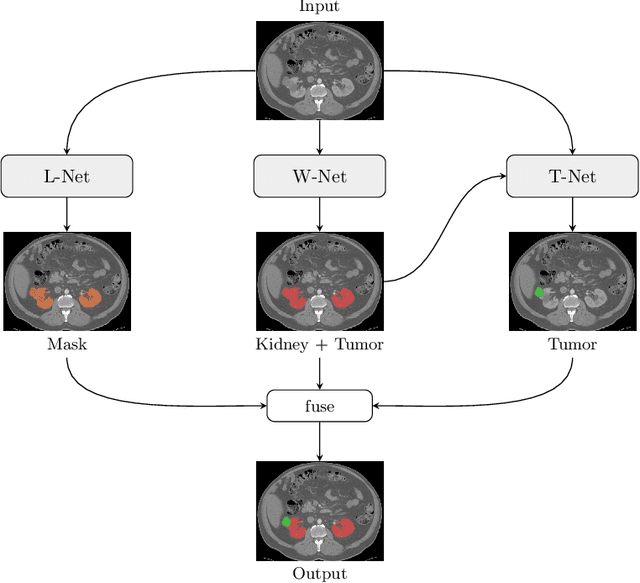
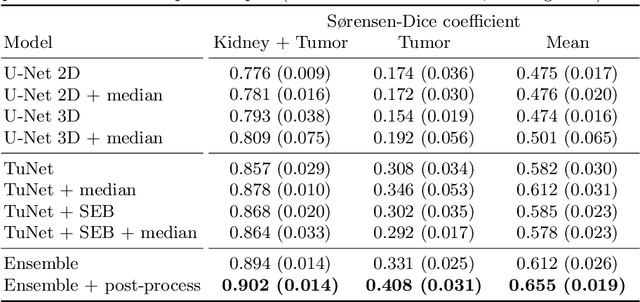
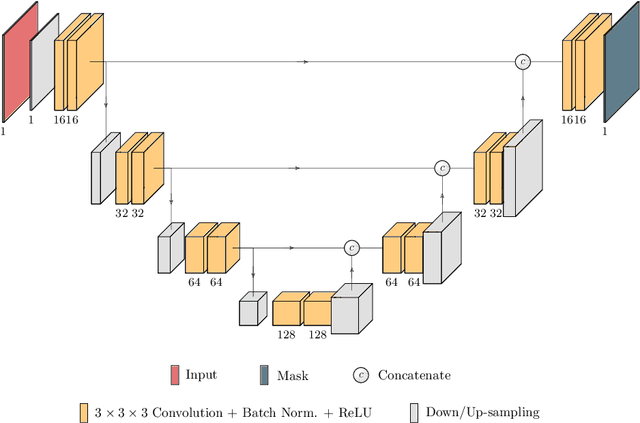
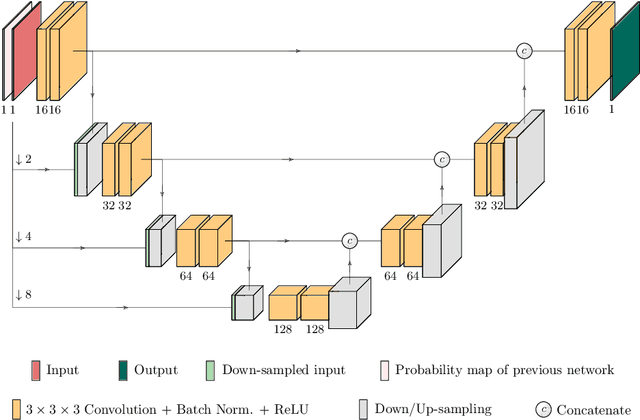
Abstract:Kidney tumor segmentation emerges as a new frontier of computer vision in medical imaging. This is partly due to its challenging manual annotation and great medical impact. Within the scope of the Kidney Tumor Segmentation Challenge 2019, that is aiming at combined kidney and tumor segmentation, this work proposes a novel combination of 3D U-Nets---collectively denoted TuNet---utilizing the resulting kidney masks for the consecutive tumor segmentation. The proposed method achieves a S{\o}rensen-Dice coefficient score of 0.902 for the kidney, and 0.408 for the tumor segmentation, computed from a five-fold cross-validation on the 210 patients available in the data.
TuNet: End-to-end Hierarchical Brain Tumor Segmentation using Cascaded Networks
Oct 11, 2019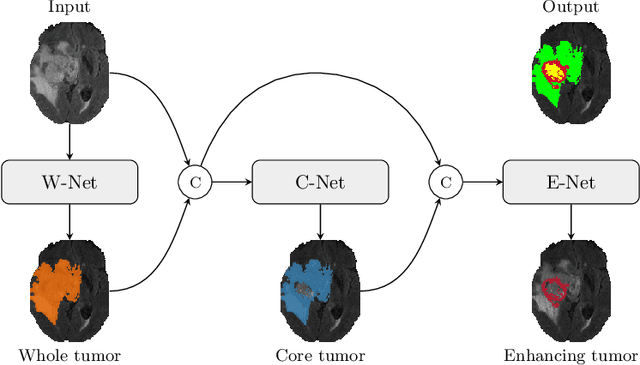
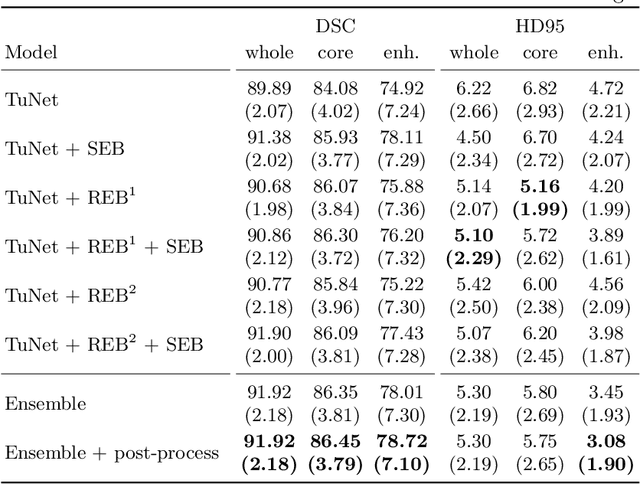
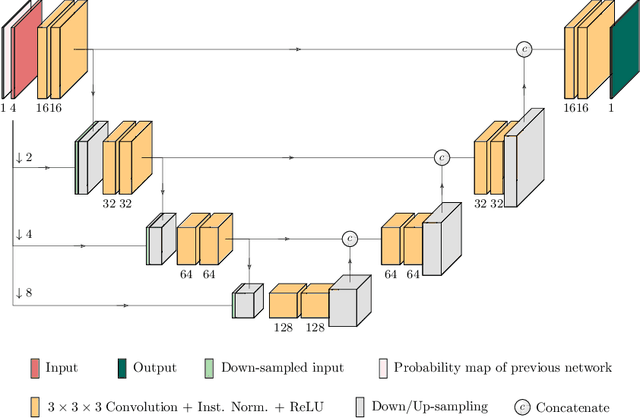
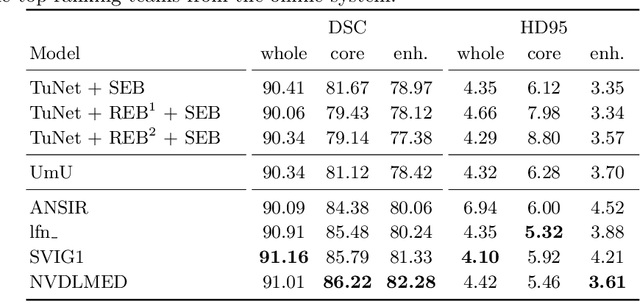
Abstract:Glioma is one of the most common types of brain tumors arising in the glial cells in the human brain and spinal cord. In addition to the threat of death, glioma treatment is also very costly. Hence, automatic and accurate segmentation and measurement from the early stages are critical in order to prolong the survival rates of the patients and to reduce the costs of health care. In the present work, we propose a novel end-to-end cascaded network for semantic segmentation that utilizes the hierarchical structure of the tumor sub-regions with ResNet-like blocks and Squeeze-and-Excitation modules after each convolution and concatenation block. By utilizing cross-validation, an average ensemble technique, and a simple post-processing technique, we obtained dice scores of 90.34, 81.12, and 78.42 and Hausdorff Distances (95th percentile) of 4.32, 6.28, and 3.70 for the whole tumor, tumor core, and enhancing tumor, respectively, on the online validation set.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge