Mary C. Scott
Contrast transfer functions help quantify neural network out-of-distribution generalization in HRTEM
Dec 09, 2025



Abstract:Neural networks, while effective for tackling many challenging scientific tasks, are not known to perform well out-of-distribution (OOD), i.e., within domains which differ from their training data. Understanding neural network OOD generalization is paramount to their successful deployment in experimental workflows, especially when ground-truth knowledge about the experiment is hard to establish or experimental conditions significantly vary. With inherent access to ground-truth information and fine-grained control of underlying distributions, simulation-based data curation facilitates precise investigation of OOD generalization behavior. Here, we probe generalization with respect to imaging conditions of neural network segmentation models for high-resolution transmission electron microscopy (HRTEM) imaging of nanoparticles, training and measuring the OOD generalization of over 12,000 neural networks using synthetic data generated via random structure sampling and multislice simulation. Using the HRTEM contrast transfer function, we further develop a framework to compare information content of HRTEM datasets and quantify OOD domain shifts. We demonstrate that neural network segmentation models enjoy significant performance stability, but will smoothly and predictably worsen as imaging conditions shift from the training distribution. Lastly, we consider limitations of our approach in explaining other OOD shifts, such as of the atomic structures, and discuss complementary techniques for understanding generalization in such settings.
Generalization Across Experimental Parameters in Machine Learning Analysis of High Resolution Transmission Electron Microscopy Datasets
Jun 20, 2023Abstract:Neural networks are promising tools for high-throughput and accurate transmission electron microscopy (TEM) analysis of nanomaterials, but are known to generalize poorly on data that is "out-of-distribution" from their training data. Given the limited set of image features typically seen in high-resolution TEM imaging, it is unclear which images are considered out-of-distribution from others. Here, we investigate how the choice of metadata features in the training dataset influences neural network performance, focusing on the example task of nanoparticle segmentation. We train and validate neural networks across curated, experimentally-collected high-resolution TEM image datasets of nanoparticles under controlled imaging and material parameters, including magnification, dosage, nanoparticle diameter, and nanoparticle material. Overall, we find that our neural networks are not robust across microscope parameters, but do generalize across certain sample parameters. Additionally, data preprocessing heavily influences the generalizability of neural networks trained on nominally similar datasets. Our results highlight the need to understand how dataset features affect deployment of data-driven algorithms.
Understanding the Influence of Receptive Field and Network Complexity in Neural-Network-Guided TEM Image Analysis
Apr 08, 2022
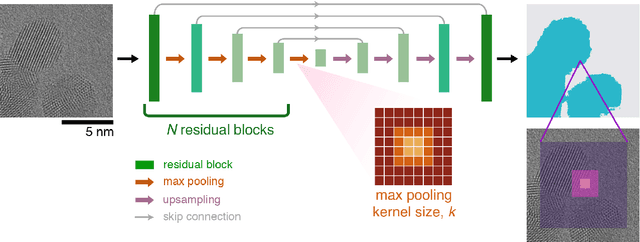

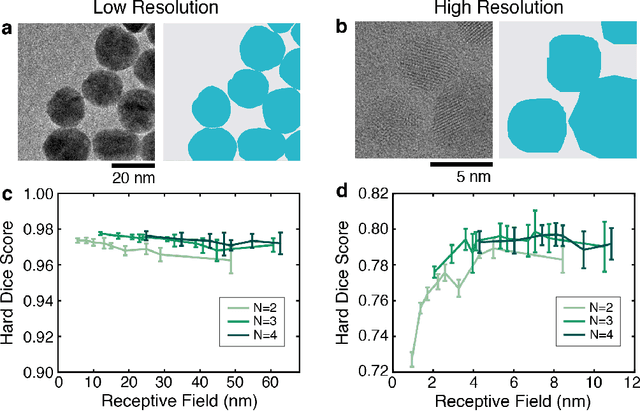
Abstract:Trained neural networks are promising tools to analyze the ever-increasing amount of scientific image data, but it is unclear how to best customize these networks for the unique features in transmission electron micrographs. Here, we systematically examine how neural network architecture choices affect how neural networks segment, or pixel-wise separate, crystalline nanoparticles from amorphous background in transmission electron microscopy (TEM) images. We focus on decoupling the influence of receptive field, or the area of the input image that contributes to the output decision, from network complexity, which dictates the number of trainable parameters. We find that for low-resolution TEM images which rely on amplitude contrast to distinguish nanoparticles from background, the receptive field does not significantly influence segmentation performance. On the other hand, for high-resolution TEM images which rely on a combination of amplitude and phase contrast changes to identify nanoparticles, receptive field is a key parameter for increased performance, especially in images with minimal amplitude contrast. Our results provide insight and guidance as to how to adapt neural networks for applications with TEM datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge