Máté E. Maros
Deep learning and abstractive summarisation for radiological reports: an empirical study for adapting the PEGASUS models' family with scarce data
Sep 18, 2025Abstract:Regardless of the rapid development of artificial intelligence, abstractive summarisation is still challenging for sensitive and data-restrictive domains like medicine. With the increasing number of imaging, the relevance of automated tools for complex medical text summarisation is expected to become highly relevant. In this paper, we investigated the adaptation via fine-tuning process of a non-domain-specific abstractive summarisation encoder-decoder model family, and gave insights to practitioners on how to avoid over- and underfitting. We used PEGASUS and PEGASUS-X, on a medium-sized radiological reports public dataset. For each model, we comprehensively evaluated two different checkpoints with varying sizes of the same training data. We monitored the models' performances with lexical and semantic metrics during the training history on the fixed-size validation set. PEGASUS exhibited different phases, which can be related to epoch-wise double-descent, or peak-drop-recovery behaviour. For PEGASUS-X, we found that using a larger checkpoint led to a performance detriment. This work highlights the challenges and risks of fine-tuning models with high expressivity when dealing with scarce training data, and lays the groundwork for future investigations into more robust fine-tuning strategies for summarisation models in specialised domains.
Simple statistical methods for unsupervised brain anomaly detection on MRI are competitive to deep learning methods
Nov 25, 2020

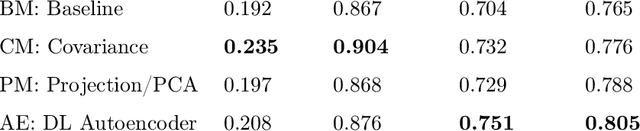

Abstract:Statistical analysis of magnetic resonance imaging (MRI) can help radiologists to detect pathologies that are otherwise likely to be missed. Deep learning (DL) has shown promise in modeling complex spatial data for brain anomaly detection. However, DL models have major deficiencies: they need large amounts of high-quality training data, are difficult to design and train and are sensitive to subtle changes in scanning protocols and hardware. Here, we show that also simple statistical methods such as voxel-wise (baseline and covariance) models and a linear projection method using spatial patterns can achieve DL-equivalent (3D convolutional autoencoder) performance in unsupervised pathology detection. All methods were trained (N=395) and compared (N=44) on a novel, expert-curated multiparametric (8 sequences) head MRI dataset of healthy and pathological cases, respectively. We show that these simple methods can be more accurate in detecting small lesions and are considerably easier to train and comprehend. The methods were quantitatively compared using AUC and average precision and evaluated qualitatively on clinical use cases comprising brain atrophy, tumors (small metastases) and movement artefacts. Our results demonstrate that while DL methods may be useful, they should show a sufficiently large performance improvement over simpler methods to justify their usage. Thus, simple statistical methods should provide the baseline for benchmarks. Source code and trained models are available on GitHub (https://github.com/vsaase/simpleBAD).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge