Louisa Jorm
The Health Gym: Synthetic Health-Related Datasets for the Development of Reinforcement Learning Algorithms
Mar 12, 2022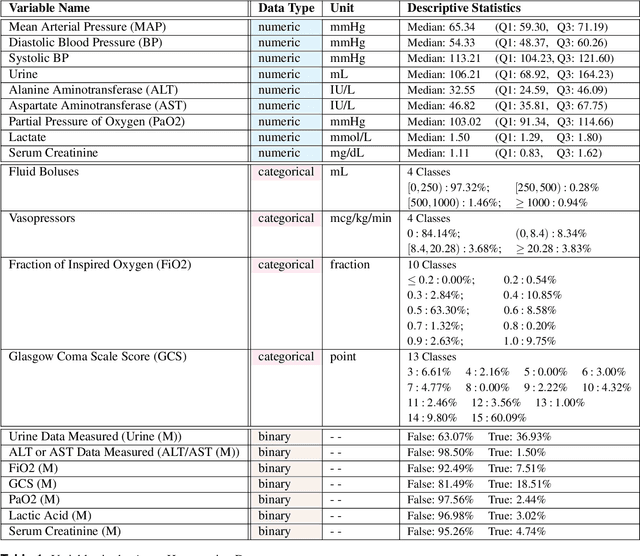
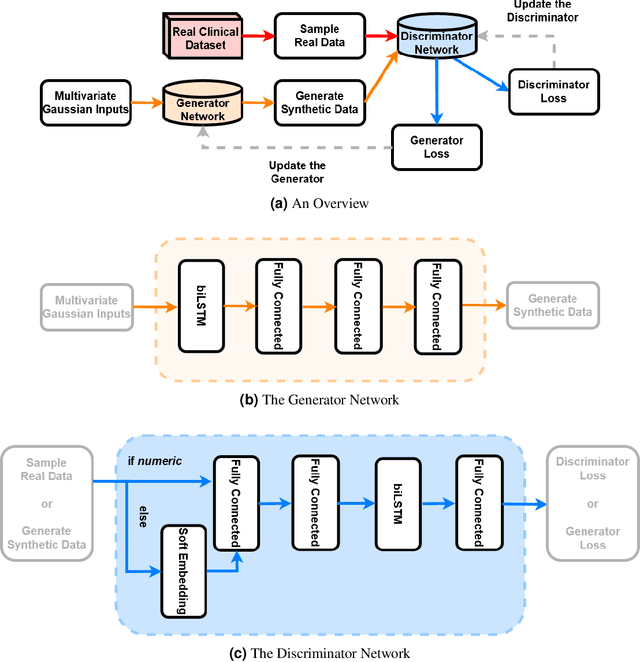
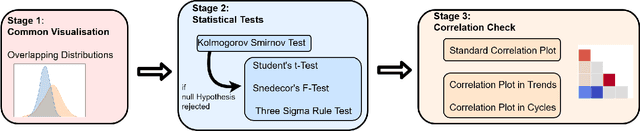
Abstract:In recent years, the machine learning research community has benefited tremendously from the availability of openly accessible benchmark datasets. Clinical data are usually not openly available due to their highly confidential nature. This has hampered the development of reproducible and generalisable machine learning applications in health care. Here we introduce the Health Gym - a growing collection of highly realistic synthetic medical datasets that can be freely accessed to prototype, evaluate, and compare machine learning algorithms, with a specific focus on reinforcement learning. The three synthetic datasets described in this paper present patient cohorts with acute hypotension and sepsis in the intensive care unit, and people with human immunodeficiency virus (HIV) receiving antiretroviral therapy in ambulatory care. The datasets were created using a novel generative adversarial network (GAN). The distributions of variables, and correlations between variables and trends over time in the synthetic datasets mirror those in the real datasets. Furthermore, the risk of sensitive information disclosure associated with the public distribution of the synthetic datasets is estimated to be very low.
Synthetic Acute Hypotension and Sepsis Datasets Based on MIMIC-III and Published as Part of the Health Gym Project
Dec 07, 2021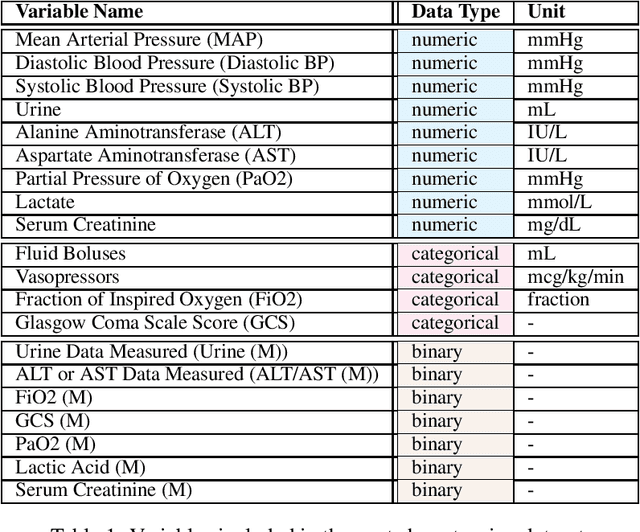

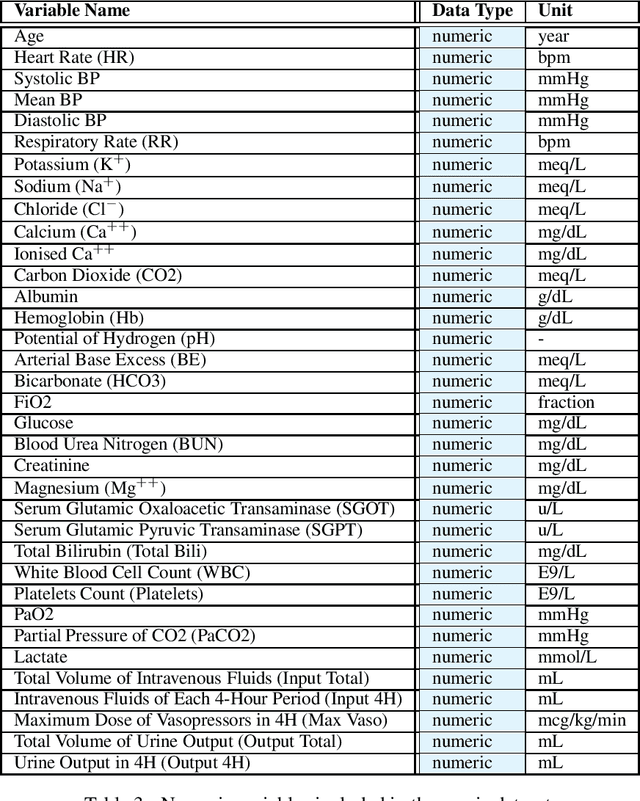
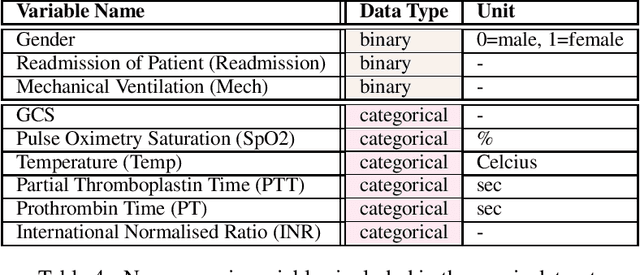
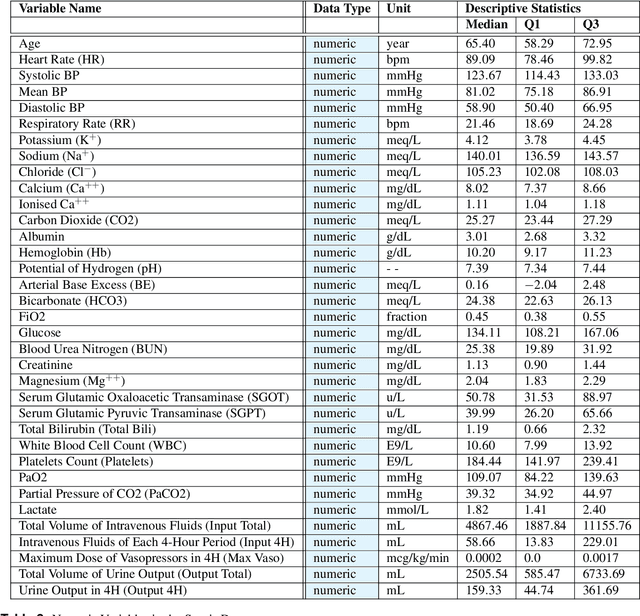
Abstract:These two synthetic datasets comprise vital signs, laboratory test results, administered fluid boluses and vasopressors for 3,910 patients with acute hypotension and for 2,164 patients with sepsis in the Intensive Care Unit (ICU). The patient cohorts were built using previously published inclusion and exclusion criteria and the data were created using Generative Adversarial Networks (GANs) and the MIMIC-III Clinical Database. The risk of identity disclosure associated with the release of these data was estimated to be very low (0.045%). The datasets were generated and published as part of the Health Gym, a project aiming to publicly distribute synthetic longitudinal health data for developing machine learning algorithms (with a particular focus on offline reinforcement learning) and for educational purposes.
De-identifying Hospital Discharge Summaries: An End-to-End Framework using Ensemble of De-Identifiers
Jan 01, 2021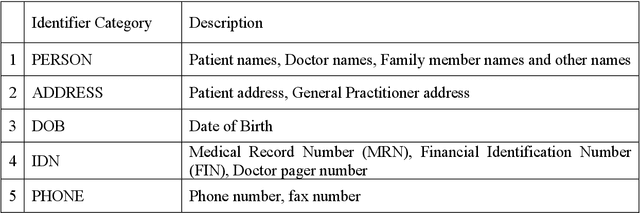
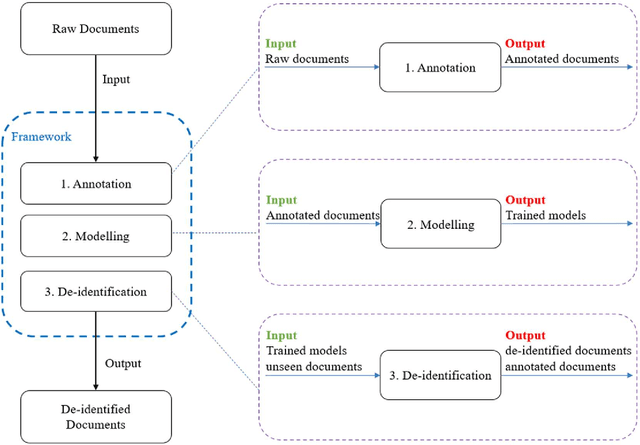
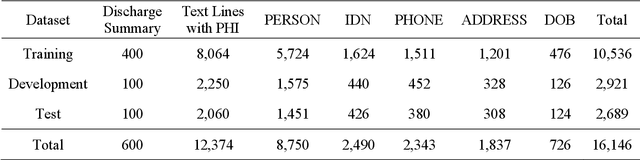
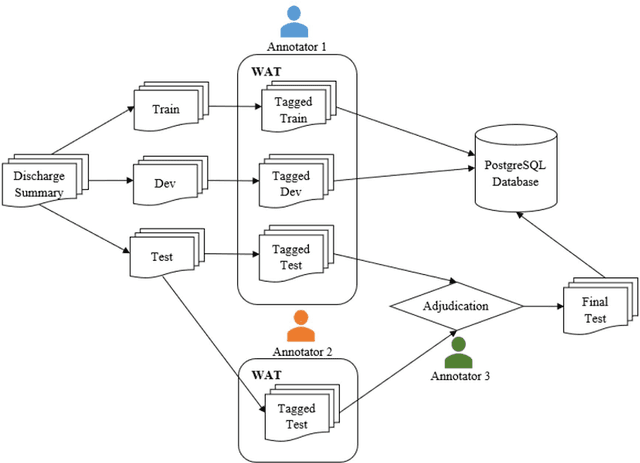
Abstract:Objective:Electronic Medical Records (EMRs) contain clinical narrative text that is of great potential value to medical researchers. However, this information is mixed with Protected Health Information (PHI) that presents risks to patient and clinician confidentiality. This paper presents an end-to-end de-identification framework to automatically remove PHI from hospital discharge summaries. Materials and Methods:Our corpus included 600 hospital discharge summaries which were extracted from the EMRs of two principal referral hospitals in Sydney, Australia. Our end-to-end de-identification framework consists of three components: 1) Annotation: labelling of PHI in the 600 hospital discharge summaries using five pre-defined categories: person, address, date of birth, individual identification number, phone/fax number; 2) Modelling: training and evaluating ensembles of named entity recognition (NER) models through the use of three natural language processing (NLP) toolkits (Stanza, FLAIR and spaCy) and both balanced and imbalanced datasets; and 3) De-identification: removing PHI from the hospital discharge summaries. Results:The final model in our framework was an ensemble which combined six single models using both balanced and imbalanced datasets for training majority voting. It achieved 0.9866 precision, 0.9862 recall and 0.9864 F1 scores. The majority of false positives and false negatives were related to the person category. Discussion:Our study showed that the ensemble of different models which were trained using three different NLP toolkits upon balanced and imbalanced datasets can achieve good results even with a relatively small corpus. Conclusion:Our end-to-end framework provides a robust solution to de-identifying clinical narrative corpuses safely. It can be easily applied to any kind of clinical narrative documents.
Predicting cardiovascular risk from national administrative databases using a combined survival analysis and deep learning approach
Nov 28, 2020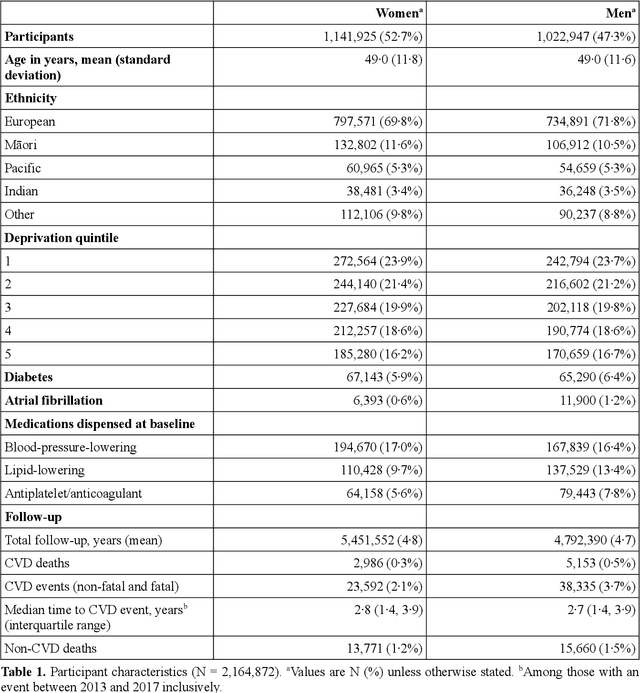
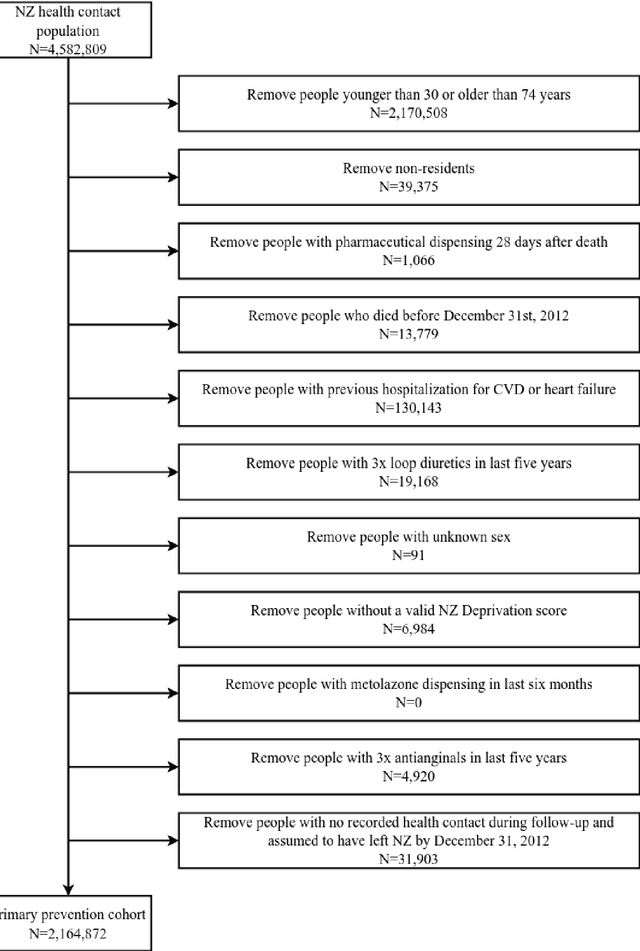
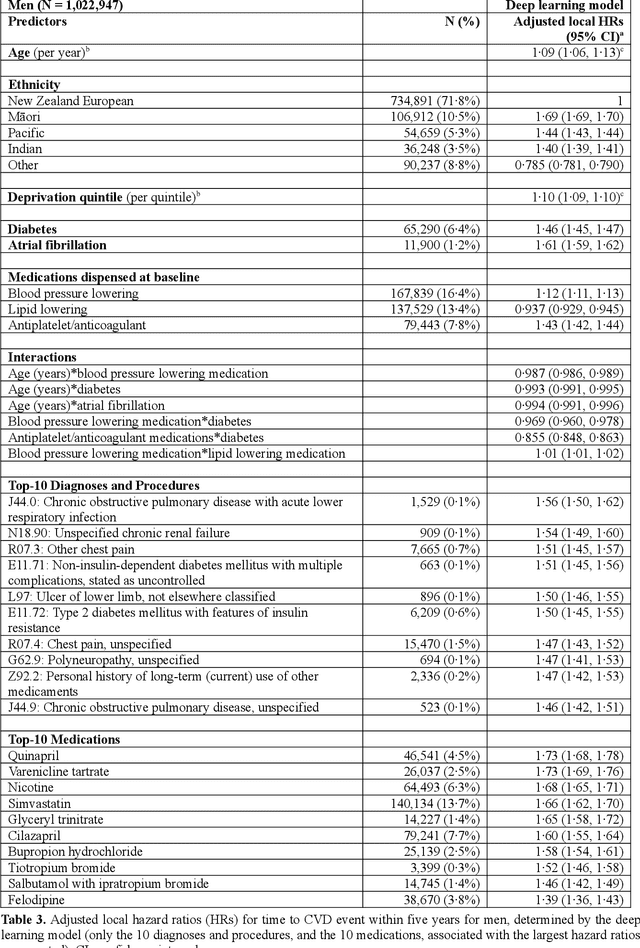
Abstract:AIMS. This study compared the performance of deep learning extensions of survival analysis models with traditional Cox proportional hazards (CPH) models for deriving cardiovascular disease (CVD) risk prediction equations in national health administrative datasets. METHODS. Using individual person linkage of multiple administrative datasets, we constructed a cohort of all New Zealand residents aged 30-74 years who interacted with publicly funded health services during 2012, and identified hospitalisations and deaths from CVD over five years of follow-up. After excluding people with prior CVD or heart failure, sex-specific deep learning and CPH models were developed to estimate the risk of fatal or non-fatal CVD events within five years. The proportion of explained time-to-event occurrence, calibration, and discrimination were compared between models across the whole study population and in specific risk groups. FINDINGS. First CVD events occurred in 61,927 of 2,164,872 people. Among diagnoses and procedures, the largest 'local' hazard ratios were associated by the deep learning models with tobacco use in women (2.04, 95%CI: 1.99-2.10) and with chronic obstructive pulmonary disease with acute lower respiratory infection in men (1.56, 95%CI: 1.50-1.62). Other identified predictors (e.g. hypertension, chest pain, diabetes) aligned with current knowledge about CVD risk predictors. The deep learning models significantly outperformed the CPH models on the basis of proportion of explained time-to-event occurrence (Royston and Sauerbrei's R-squared: 0.468 vs. 0.425 in women and 0.383 vs. 0.348 in men), calibration, and discrimination (all p<0.0001). INTERPRETATION. Deep learning extensions of survival analysis models can be applied to large health administrative databases to derive interpretable CVD risk prediction equations that are more accurate than traditional CPH models.
A Deep Representation of Longitudinal EMR Data Used for Predicting Readmission to the ICU and Describing Patients-at-Risk
May 21, 2019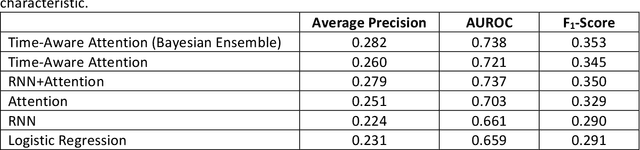
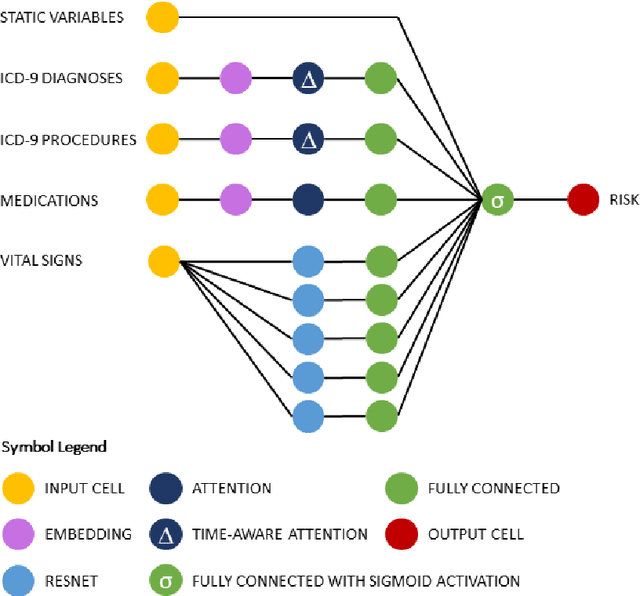
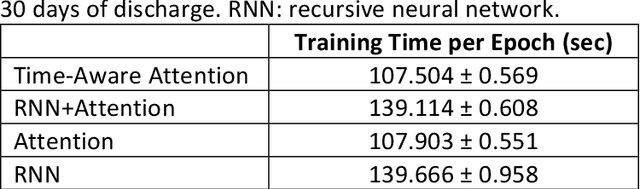
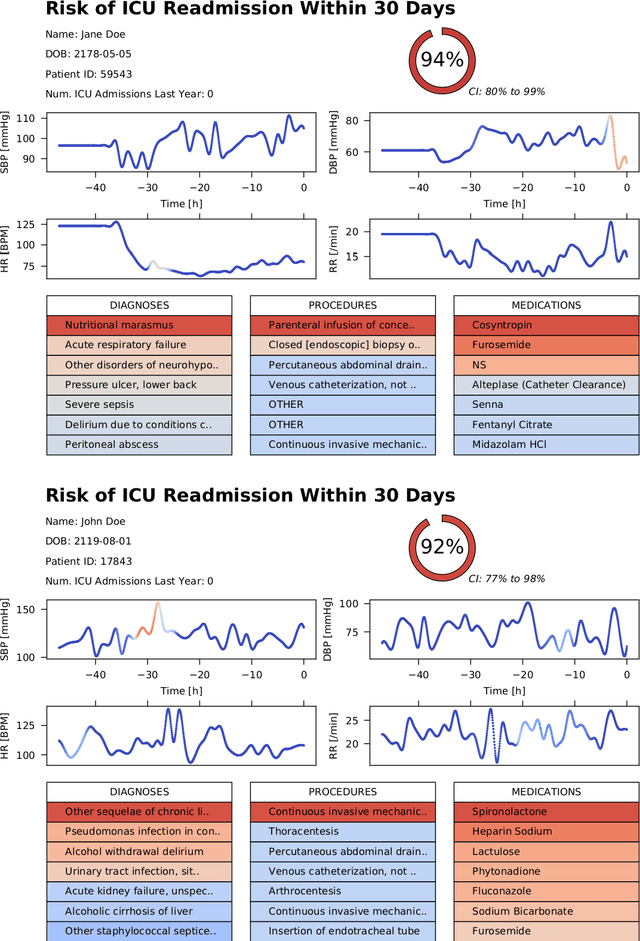
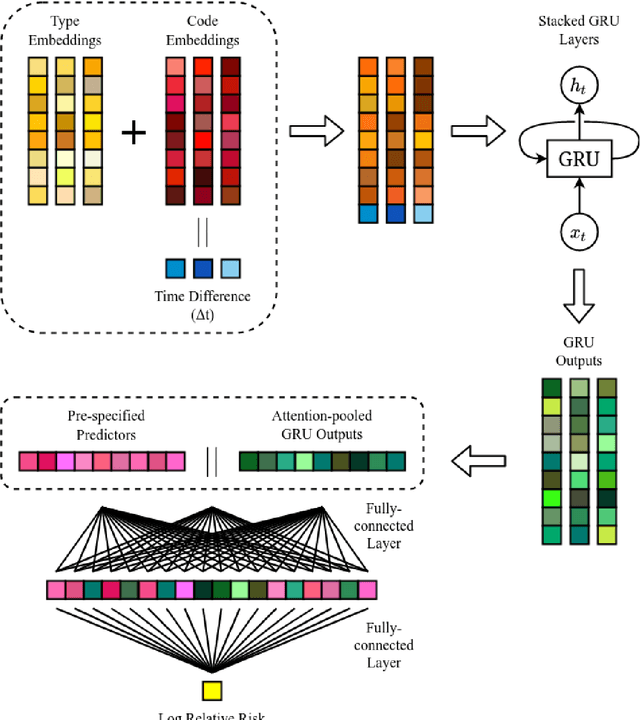
Abstract:Objective: To evaluate the feasibility of using an attention-based neural network for predicting the risk of readmission within 30 days of discharge from the intensive care unit (ICU) based on longitudinal electronic medical record (EMR) data and to leverage the interpretability of the model to describe patients-at-risk. Methods: A "time-aware attention" model was trained using publicly available EMR data (MIMIC-III) associated with 45,298 ICU stays for 33,150 patients. The analysed EMR data included static (patient demographics) and timestamped variables (diagnoses, procedures, medications, and vital signs). Bayesian inference was used to compute the posterior distribution of network weights. The prediction accuracy of the proposed model was compared with several baseline models and evaluated based on average precision, AUROC, and F1-Score. Odds ratios (ORs) associated with an increased risk of readmission were computed for static variables. Diagnoses, procedures, and medications were ranked according to the associated risk of readmission. The model was also used to generate reports with predicted risk (and associated uncertainty) justified by specific diagnoses, procedures, medications, and vital signs. Results: A Bayesian ensemble of 10 time-aware attention models led to the highest predictive accuracy (average precision: 0.282, AUROC: 0.738, F1-Score: 0.353). Male gender, number of recent admissions, age, admission location, insurance type, and ethnicity were all associated with risk of readmission. A longer length of stay in the ICU was found to reduce the risk of readmission (OR: 0.909, 95% credible interval: 0.902, 0.916). Groups of patients at risk included those requiring cardiovascular or ventilatory support, those with poor nutritional state, and those for whom standard medical care was not suitable, e.g. due to contraindications to surgery or medications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge